Figure 2. Genome-wide screening with differential expression and H3K27me3 levels significantly enriches for transdifferentiation factors.
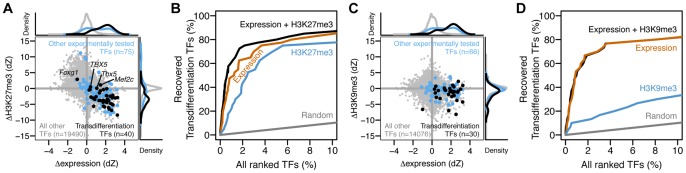
(A) We compared six human and eight mouse pairs of cell types with known transdifferentiation factors (Table 1) and plotted the differential expression (x-axis) and differential H3K27me3 modification (y-axis) for these factors (black points) as well as all other TFs in the genome (grey points). We also indicated (blue points) the subset of these other TFs that were experimentally tested for their ability to convert cell types, but not included in the conversion protocols (Table S2). Each point represents 1 gene in 1 pair of cell types. Most transdifferentiation factors are more highly expressed in the target cell (right side of the y-axis) and more highly H3K27me3 modified in the source cell (below the x-axis). The marginal distributions depict the differential expression (top) and modification (right) of transdifferentiation factors (black curve) compared to all other transcription factors (grey curve), as well as the subset that were experimentally tested (blue curve). (B) We ranked all transcription factors by differential expression (orange), differential H3K27me3 modification (blue), or a combination of both (black) and then calculated the percentage of tested transdifferentiation factors (y-axis) as a function of the position in the ranked list. (C) Plot similar to (A) showing differential H3K9me3 levels in place of H3K27me3. In contrast to H3K27me3 levels, transdifferentiation factors exhibit on average only a slight differential H3K9me3 modification. Six human and four mouse pairs of cell types were used in this analysis, as H3K9me3 modification profiles were not available for three mouse tissues (myoblast, heart, neuron; Table 1). (D) H3K9me3 modification provides minimal information beyond expression for identifying transdifferentiation factors. Data sources are listed in Table S1.
