Five out of 11 related transcription factors were found to mediate the cytokinin response based on complementation analysis of a cytokinin-signaling mutant.
Abstract
Cytokinins play critical roles in plant growth and development, with the transcriptional response to cytokinin being mediated by the type-B response regulators. In Arabidopsis (Arabidopsis thaliana), type-B response regulators (ARABIDOPSIS RESPONSE REGULATORS [ARRs]) form three subfamilies based on phylogenic analysis, with subfamily 1 having seven members and subfamilies 2 and 3 each having two members. Cytokinin responses are predominantly mediated by subfamily 1 members, with cytokinin-mediated effects on root growth and root meristem size correlating with type-B ARR expression levels. To determine which type-B ARRs can functionally substitute for the subfamily 1 members ARR1 or ARR12, we expressed different type-B ARRs from the ARR1 promoter and assayed their ability to rescue arr1 arr12 double mutant phenotypes. ARR1, as well as a subset of other subfamily 1 type-B ARRs, restore the cytokinin sensitivity to arr1 arr12. Expression of ARR10 from the ARR1 promoter results in cytokinin hypersensitivity and enhances shoot regeneration from callus tissue, correlating with enhanced stability of the ARR10 protein compared with the ARR1 protein. Examination of transfer DNA insertion mutants in subfamilies 2 and 3 revealed little effect on several well-characterized cytokinin responses. However, a member of subfamily 2, ARR21, restores cytokinin sensitivity to arr1 arr12 roots when expressed from the ARR1 promoter, indicating functional conservation of this divergent family member. Our results indicate that the type-B ARRs have diverged in function, such that some, but not all, can complement the arr1 arr12 mutant. In addition, our results indicate that type-B ARR expression profiles in the plant, along with posttranscriptional regulation, play significant roles in modulating their contribution to cytokinin signaling.
Cytokinins are phytohormones that play critical roles in plant growth and development, including regulation of cell division and metabolism, stimulation of chloroplast development, modulation of shoot and root development, and delay of leaf senescence (Mok, 1994; Haberer and Kieber, 2002; Kakimoto, 2003). Cytokinin signal transduction is mediated by a multistep phosphorelay that involves cytokinin receptors, phosphotransfer proteins, and type-B response regulators (Kakimoto, 2003; To and Kieber, 2008; Werner and Schmülling, 2009). These relay the cytokinin signal from the membrane to the nucleus, where the type-B response regulators induce the transcription of many genes. In Arabidopsis (Arabidopsis thaliana), there are three cytokinin receptors (ARABIDOPSIS HIS KINASE2 [AHK2], AHK3, and AHK4; Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001; Kakimoto, 2003), five phosphotransfer proteins (ARABIDOPSIS HIS-CONTAINING PHOSPHOTRANSFER PROTEINS; Hwang and Sheen, 2001; Hutchison et al., 2006), and 11 type-B response regulators (ARABIDOPSIS RESPONSE REGULATORS [ARRs]; Sakai et al., 2001; Mason et al., 2005). Genetic analysis has demonstrated roles for each of these families in cytokinin-mediated processes (Mähönen et al., 2000, 2006; Higuchi et al., 2004; Nishimura et al., 2004; To et al., 2004; Mason et al., 2005; Hutchison et al., 2006; Yokoyama et al., 2007; Argyros et al., 2008).
According to this model, the type-B ARRs play a pivotal role in the early transcriptional response of plants to cytokinin. The type-B ARRs are structurally related, each possessing a receiver domain that is phosphorylated on a conserved Asp residue, as well as a long C-terminal extension with a Myb-like DNA-binding domain (Imamura et al., 1999; Hosoda et al., 2002). The ability of the Myb-like domain of type-B ARRs to bind DNA has been demonstrated in several studies (Sakai et al., 2000; Hosoda K, et al., 2002), and multiple lines of evidence support a role of type-B ARRs as transcription factors (Sakai et al., 2000, 2001; Imamura et al., 2001, 2003; Lohrmann et al., 2001; Hosoda et al., 2002; Mason et al., 2004, 2005; Rashotte et al., 2006; Liang et al., 2012; Tsai et al., 2012). Elimination of three type-B ARRs, ARR1, ARR10, and ARR12, severely curtails the ability of cytokinin to induce changes in gene expression, demonstrating the importance of the type-B ARRs in the initial cytokinin signal transduction pathway and indicating that the type-B ARRs act at the top of a transcriptional cascade (Argyros et al., 2008; Ishida et al., 2008).
The 11 type-B ARRs of Arabidopsis fall into three subfamilies based on phylogenetic analysis, with subfamily 1 containing seven members and subfamilies 2 and 3 each containing two members (Mason et al., 2004). The members of subfamily 1 have been most extensively characterized. The type-B ARRs of subfamily 1 have the broadest expression pattern in Arabidopsis, and genetic analysis indicates that at least five members, ARR1, ARR2, ARR10, ARR11, and ARR12, of the subfamily mediate cytokinin signaling (Mason et al., 2005; Yokoyama et al., 2007; Argyros et al., 2008; Ishida et al., 2008). In this study, we describe results obtained from two approaches to characterize the roles of type-B ARRs in cytokinin signaling. First, we assessed the function of all 11 type-B ARRs under the same expression context based on their ability to complement the arr1 arr12 mutant when driven from the ARR1 promoter. Second, we examined the effect of disruption of type-B ARRs from subfamilies 2 and 3. Results from these studies indicate that the type-B ARRs have diverged in function, such that some, but not all, complement arr1 arr12. In addition, our results indicate that type-B ARR expression profiles in the plant, along with posttranscriptional regulation, may play significant roles in modulating their contribution to cytokinin signaling.
RESULTS
Expression and the Contribution of Type-B ARRs to Root Growth
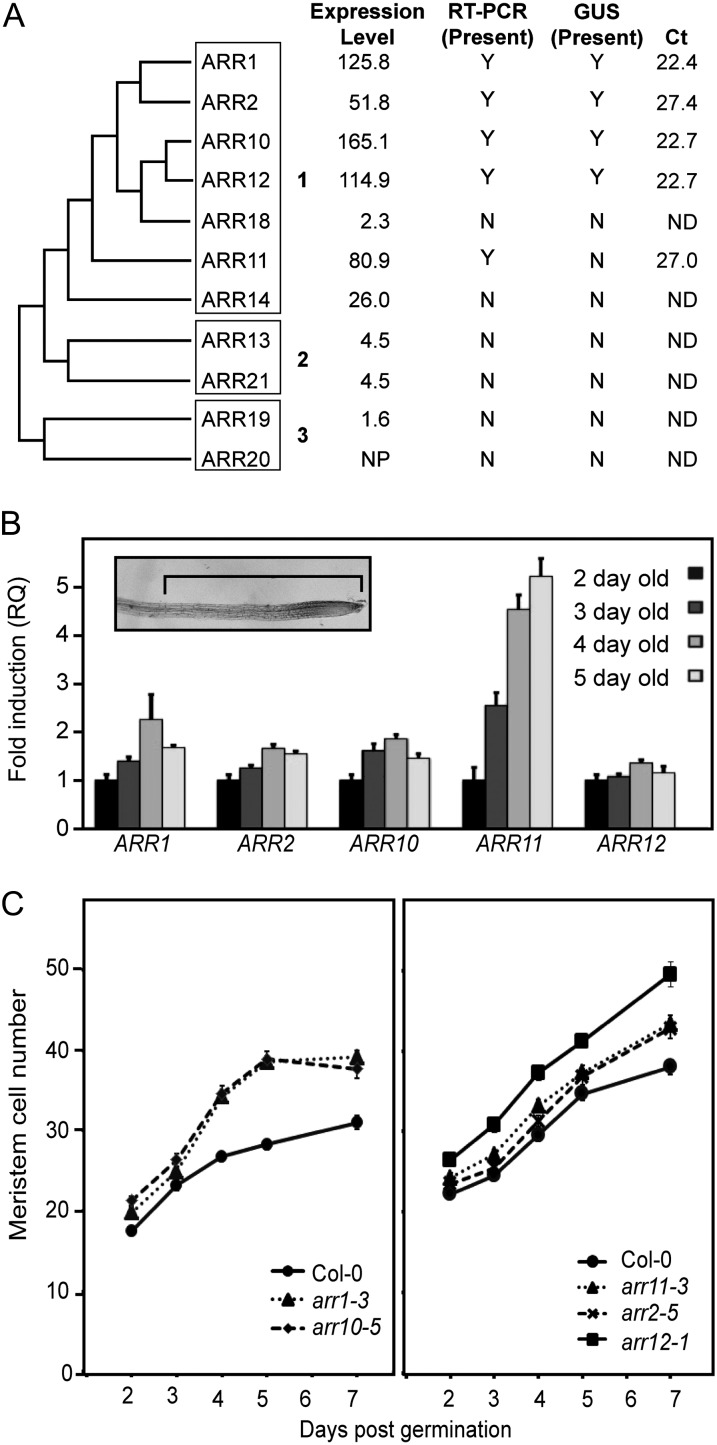
In Arabidopsis, there are 11 type-B ARRs that are divided into three subfamilies based on sequence homology (Fig. 1A; Mason et al., 2004). Data from microarray studies, semiquantitative reverse transcription (RT)-PCR, and GUS reporter analysis indicate that subfamily 1 members ARR1, ARR2, ARR10, ARR11, and ARR12 are the most highly expressed type-B ARRs in the roots (Fig. 1A; Birnbaum et al., 2003; Imamura et al., 2003; Mason et al., 2004; Tajima et al., 2004; Schmid et al., 2005). Genetic studies suggest that ARR1, ARR10, and ARR12 are the primary components of the cytokinin response in the root (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008). To gain information about temporal regulation of expression for the five family members we could detect by PCR-based techniques, we performed quantitative RT-PCR on RNA isolated from root tips of seedlings 2, 3, 4, and 5 d after germination (Fig. 1B). The region of the root used for our analysis includes the stem cell niche, the cell division zone, the transition zone, and the initial part of the elongation/differentiation zone (Dello Ioio et al., 2008a). Expression of ARR12 remained relatively consistent during this time period (Fig. 1B). At the other extreme, ARR11 exhibited a 5-fold increase in expression between days 2 and 5. ARR1, ARR2, and ARR10 all exhibited some increase in expression between days 2 and 4, with ARR1 expression increasing 2-fold during this time period (Fig. 1B). Overall, based on average threshold cycle (Ct) values obtained from quantitative RT-PCR (Fig. 1A), the expression levels of ARR2 and ARR11 are substantially less than those of ARR1, ARR10, and ARR12, even at their time point of maximal expression.
Figure 1.
Expression of type-B ARRs in Arabidopsis roots varies during early stages of growth and correlates with effects on root meristem size. A, Expression of type-B ARRs based on microarray analysis, RT-PCR, GUS fusion analysis, and quantitative RT-PCR. A cladogram based on the receiver domains of subfamily 1, 2, and 3 type-B ARRs was constructed using the phylogeny.fr pipeline (Dereeper et al., 2008). Absolute expression level in 17-d-old roots is derived from the microarray data of Schmid et al. (2005), as accessed through the Arabidopsis eFP Browser (Winter et al., 2007), with the housekeeping gene β-TUBULIN3 (At5g62700) having an expression level of 1264.4 by way of comparison. The presence (Y) or absence (N) of the type-B ARRs in roots based on RT-PCR and translational GUS fusions is from Mason et al. (2004). Average Ct values are from the point of maximal expression based on quantitative RT-PCR analysis in root tips, with a lower Ct value indicating higher expression. NP, Gene was not represented on the array; ND, not determined. B, ARR1, ARR2, ARR10, ARR11, and ARR12 transcript levels in root tips 2, 3, 4, and 5 d after germination. Transcript levels are expressed relative to day 2. Inset image shows a 5-d-old root tip (tip on right) with the 1-mm region used for isolation of RNA indicated. C, Effect of individual arr mutations on meristem size. Meristem size was determined by counting cell number as described (Dello Ioio et al., 2008b). Error bars represent se. Significant differences from the wild type (Bonferroni-corrected comparison of statistical difference, P < 0.05) are found for arr1 (days 4–7), arr2 (days 4 and 7), arr10 (days 2 and 4–7), arr11 (days 5–7), and arr12 (days 2–7).
To determine if temporal expression patterns of these type-B ARRs correlated with their role in root development, we examined the effect of single type-B ARR mutants on root meristem size (Fig. 1C). Root meristem size was determined by counting the number of meristematic cells at days 2 through 7 after germination. The arr12-1 mutant exhibited an enlarged meristem throughout this time period, whereas the arr1-3 mutant did not exhibit a strong effect until day 4 (Fig. 1C), which is consistent with previous reports (Dello Ioio et al., 2008b; Moubayidin et al., 2010). The arr10-5 mutant behaved similarly to the arr1-3 mutant, also showing little effect early after germination but a more pronounced effect at day 4 and thereafter. The arr2-5 and arr11-3 mutants had only a weak effect on meristem size, with their contribution most apparent later. Thus, overall, the effects of the individual type-B ARRs on meristem size are consistent with (1) their absolute expression level and (2) temporal changes in their expression level.
Functional Analysis of Subfamily 1 Type-B ARRs in Arabidopsis
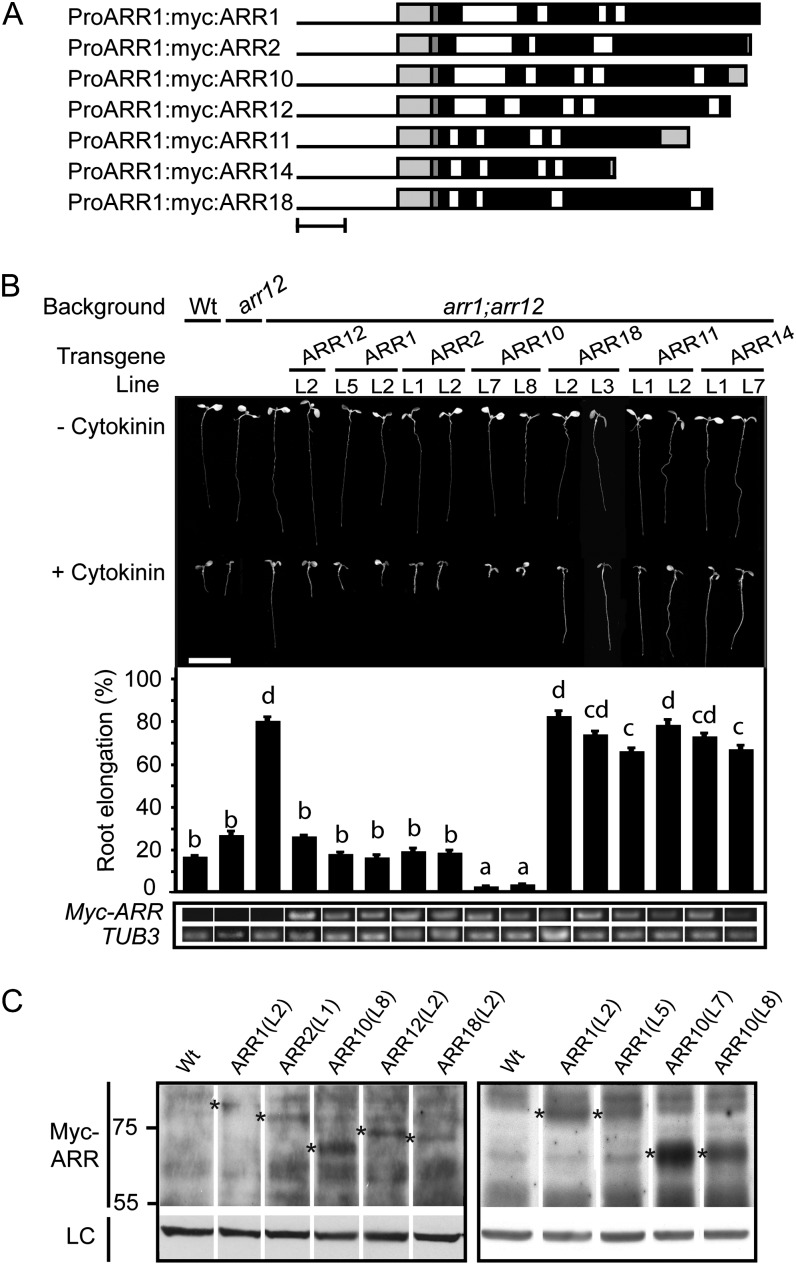
The differing expression patterns of the type-B ARRs raised the question as to whether the function of these proteins is interchangeable. To address this question, we took advantage of the partial cytokinin insensitivity (hyposensitivity) of the arr1 arr12 double mutant (Mason et al., 2005; Argyros et al., 2008) to determine which type-B ARRs could functionally substitute for activity of ARR1 (or ARR12, as this mutant-based assay is not unequivocal for ARR1). We expressed different members of subfamily 1 from the ARR1 promoter (Fig. 2A), incorporating a Myc epitope tag into the transgene to facilitate detection and comparison of transgene expression. To minimize potential adverse effects of a tag on function, only a single 10-amino acid Myc epitope was used, and the tag was incorporated at an analogous position at the amino termini of each encoded protein, proximate to the receiver domain. This type of functional analysis has been used before, notably to examine the function of the ethylene receptor family in plant growth (Wang et al., 2003), and circumvents artifacts that can arise due to ectopic overexpression, such as that driven by the Cauliflower mosaic virus 35S promoter.
Figure 2.
A subset of subfamily 1 ARRs functionally complement the root growth phenotype of the arr1 arr12 mutant. A, Schematic of the subfamily 1 constructs used in this study. Bar = 500 nucleotides. Black line indicates ARR1 promoter, light-gray box on left side indicates ARR1 5′-UTR, dark-gray box indicates myc sequence, black boxes indicate exons, white boxes indicate introns, and light-gray box on right side indicates 3′-UTR. B, Root growth inhibition by 1 µm BA. The top portion shows representative 7-d-old seedlings grown in presence or absence of 1 µm BA. The middle portion shows the root elongation response of seedlings grown on media containing 1 μm BA expressed as a percentage of root growth of siblings grown on dimethyl sulfoxide (DMSO) control media. Root growth from day 4 through day 7 was measured. Lines were analyzed for significant differences in their responsiveness to cytokinin based on Tukey’s multiple range test among the means on the ANOVA (P < 0.05). Lines designated with the same letter exhibit no significant difference. The bottom portion shows transcript levels of the ARR transgenes in the roots of 7-d-old seedlings, based on RT-PCR from the common sequence involving the ARR1 5′-UTR and the c-Myc epitope tag. β-tubulin3 (At5g62700) was used as a loading control. Amplicons are of the same exposure. Error bars represent se. Bar = 1 cm. C, Protein levels of selected ARR transgenes based on immunoblot analysis using the Myc epitope tag. Asterisks indicate full-length transgenic protein. Hsp70 protein was immunologically detected as a loading control (LC). Predicted molecular masses are 76.4 kD (ARR1), 73.8 kD (ARR2), 62.9 kD (ARR10), 66.8 kD (ARR12), and 69.7 kD (ARR18).
Multiple independent transgenic lines were assayed for their ability to functionally complement cytokinin hyposensitivity of the arr1 arr12 mutant. We found that ARR1 (6/6 lines), ARR2 (6/6 lines), ARR10 (9/11 lines), and ARR12 (11/14 lines) but not ARR11 (0/11 lines), ARR14 (0/16 lines), or ARR18 (0/14 lines) could restore cytokinin sensitivity to the arr1 arr12 mutant in root growth assays. Data for a subset of these lines is shown in Figure 2B, with a line capable of rescue being included if any such was observed. We included the arr12 mutant in this analysis because it contains wild-type ARR1 and thus represents the level of response one might anticipate if transgenic ARR1 were expressed in arr1 arr12 under completely native conditions. We defined complete complementation of arr1 arr12 as a recovery to wild-type sensitivity or better, with a response that is significantly different from that of arr1 arr12 (P < 0.05). We defined partial complementation as a recovery to at least 25% of the wild-type sensitivity, with a response that is significantly different from that of arr1 arr12 (P < 0.05). In the absence of cytokinin, wild-type, arr12, arr1 arr12, and the transgenic lines are all of similar appearance, but significant differences can be observed in their root growth response to 1 µm benzyladenine (BA; Fig. 2B). Transgenic expression of ARR1, ARR2, ARR10, and ARR12 all reverted the cytokinin insensitivity of arr1 arr12 to wild-type levels or better (i.e. a complete complementation of the mutant phenotype). By contrast, transgenic expression of ARR11, ARR14, and ARR18 failed to complement the mutant phenotype, although a slight but statistically significant increase in cytokinin sensitivity was noted for one line each of ARR11 (line 1) and ARR14 (line 7).
This same pattern of complementation was also observed in hypocotyl elongation assays, where cytokinin normally acts to inhibit hypocotyl growth in dark-grown seedlings (Supplemental Fig. S1). Hypocotyl length was similar for all lines in the absence of cytokinin, but in the presence of cytokinin, the transgenic lines of ARR1, ARR2, ARR10, and ARR12 were all capable of partially or completely reverting the cytokinin insensitivity of arr1 arr12 to a wild-type level of sensitivity. The same pattern of complementation was also observed in the ability of the transgene to rescue the enlarged seed size phenotype observed in the arr1 arr12 mutant (Supplemental Fig. S1). The inability of ARR11, ARR14, and ARR18 to rescue the arr1 arr12 mutant is not due to poor transgene expression, as their expression was similar to or higher than other family members that rescued the arr1 arr12 phenotypes (Fig. 2B; Supplemental Fig. S2). We were also able to confirm protein accumulation for several of these transgenic proteins (Fig. 2C). We could consistently detect the tagged version of ARR10 (predicted molecular mass of 62.9 kD), and also detected less abundant protein bands corresponding to the tagged versions of ARR1 (76.4 kD), ARR2 (73.8 kD), ARR12 (66.8 kD), and ARR18 (69.7 kD). We could not detect ARR11 (59.8 kD) or ARR14 (44.3 kD), although in many cases, type-B ARR protein levels were below our limits of detection or obscured by nonspecific background bands, even under conditions where rescue was observed. These data support a functional difference among the subfamily 1 type-B ARRs.
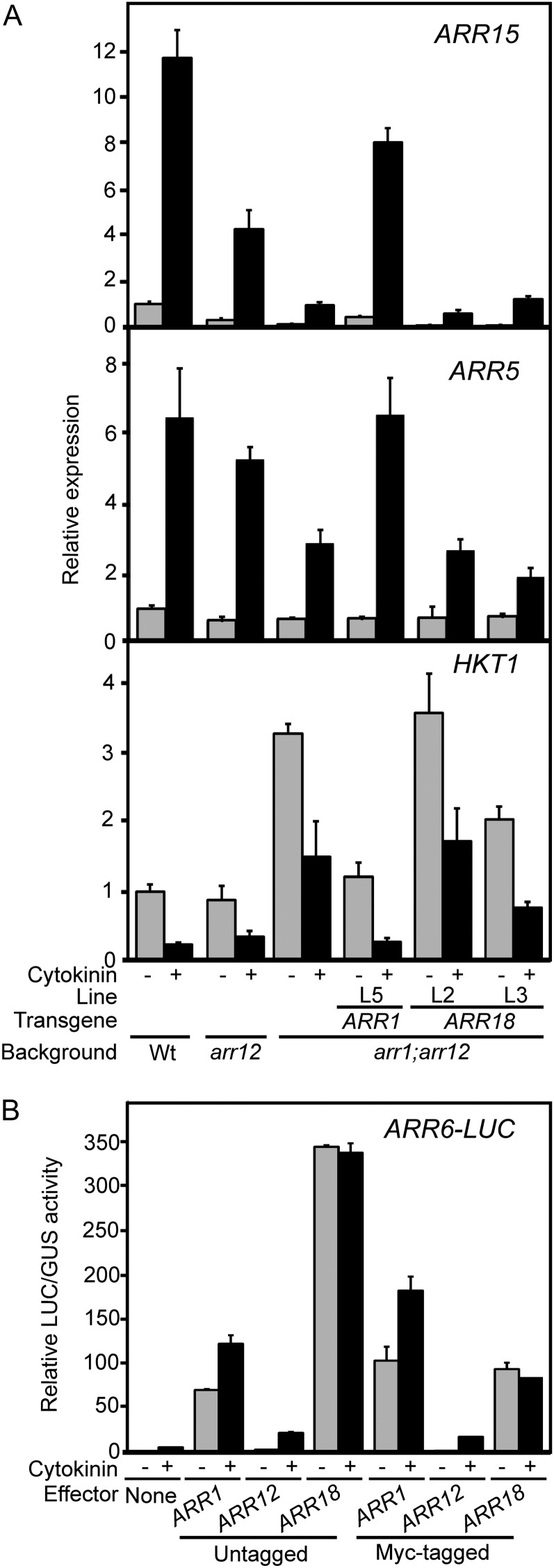
In tandem with the physiological response phenotypes, we also examined molecular responses to determine how gene regulation correlates with the ability of the transgenes to functionally complement the arr1 arr12 mutant (Fig. 3A). For this purpose, we examined the cytokinin-mediated induction of the primary-response genes ARR15 and ARR5 (Taniguchi et al., 2007; Argyros et al., 2008) and repression of HIGH-AFFINITY K+ TRANSPORTER1 (HKT1; Mason et al., 2010). ARR15 and ARR5 are induced approximately 11-fold and 7-fold, respectively, in response to 2-h cytokinin treatment in wild-type roots; however, this induction is severely attenuated in the arr1 arr12 mutant (Fig. 3A). Despite comparable RNA and protein accumulation (Fig. 2, B and C), ARR1 but not ARR18 was able to rescue this molecular phenotype for ARR15 and ARR5 expression (Fig. 3A). HKT1, a gene whose product is responsible for removing sodium ions from the root xylem, is repressed by cytokinin treatment and significantly elevated in arr1 arr12 (Fig. 3A; Mason et al., 2010). ARR1 fully rescued and one of the ARR18 lines (L3) partially rescued the molecular phenotype for HKT1 (Fig. 3A). As a complement to this molecular study, we also examined the ability of type-B ARRs to stimulate expression of a cytokinin-regulated LUCIFERASE (LUC) reporter gene in a transient protoplast expression system (Hwang and Sheen, 2001). ARR1, ARR12, and ARR18 all stimulated pARR6:LUC expression, demonstrating that all three proteins are functional transcription factors, but only ARR1 and ARR12 activated the reporter gene in a cytokinin-dependent manner (Fig. 3B). Addition of the N-terminal Myc tag appears to decrease activity of ARR18 based on the transient protoplast assay, but ARR18 activity is still comparable to that of ARR1 and is substantially above that of ARR12, both of which rescue the arr1 arr12 mutant (Fig. 3B). These results point to a fundamental difference in the ability of ARR1 and ARR18 to regulate expression of known cytokinin primary-response genes and suggests that their differing abilities to rescue the arr1 arr12 mutant may be due to such differences in gene regulation.
Figure 3.
Effect of the ARR1 and ARR18 transgenes on cytokinin-regulated expression of cytokinin primary-response genes. A, Transcript levels of ARR15, ARR5, and HKT1 were determined in the wild type (Wt), arr12, arr1 arr12, and transgenic lines of arr1 arr12 containing either proARR1:myc:ARR1 (ARR1) or proARR:myc:ARR18 (ARR18). RNA was isolated from the roots of 14-d-old seedlings treated for 2 h with 10 µm BA or a DMSO vehicle control, and the relative expression levels of ARR15, ARR5, and HKT1 were determined based on quantitative RT-PCR. Error bars indicate se. B, Functional analysis of ARR1, ARR12, and ARR18 in the Arabidopsis protoplast transient expression system. Protoplasts were cotransfected with the ARR6-LUC reporter and an effector plasmid expressing ARR1, ARR12, or ARR18, using untagged or Myc-tagged versions of the type-B ARRs. Transfection of the reporter gene with empty vector DNA were used as a control. Transfected protoplasts were treated without (–CK) or with (+CK) the cytokinin trans-zeatin (100 nm). The results are shown as the means of relative LUC activities from duplicate samples with error bars that indicate sd.
ARR10 Confers a Hypersensitivity Phenotype When Expressed in the Same Context as ARR1
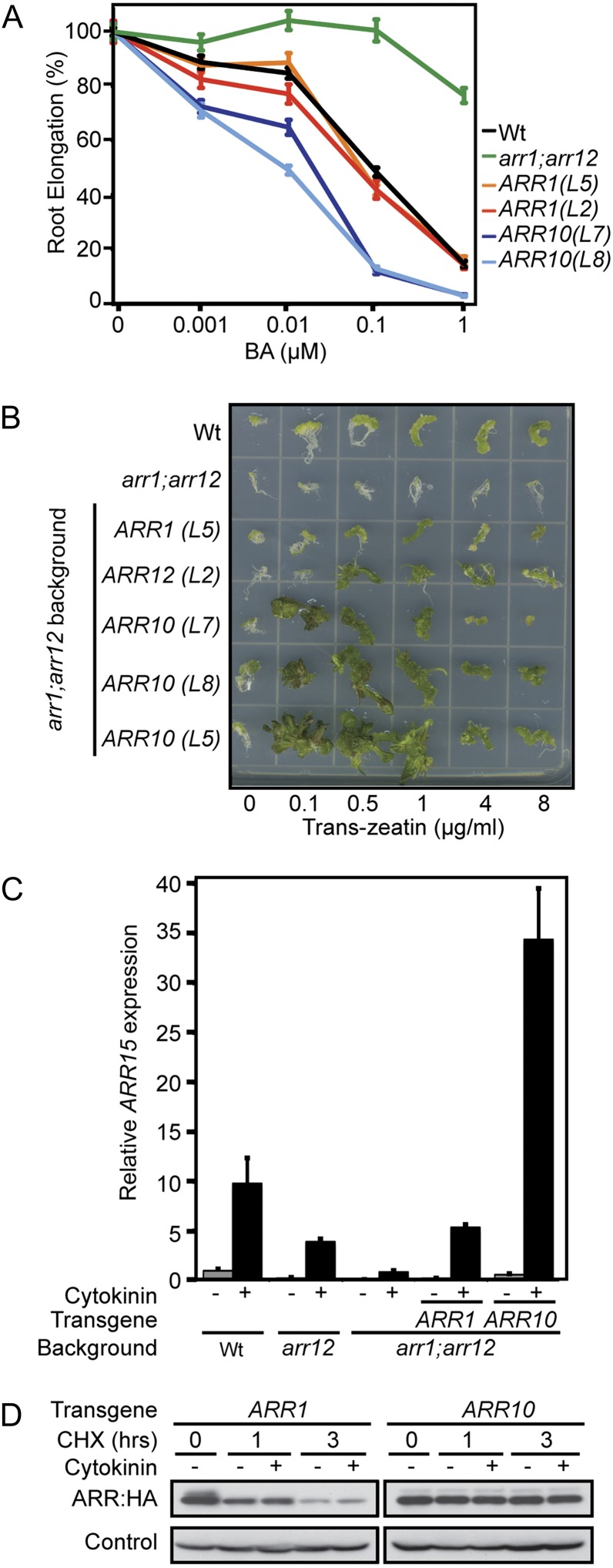
We observed that ARR10, when expressed from the ARR1 promoter, results in a cytokinin hypersensitivity phenotype in the roots, despite accumulating levels of transgene transcript comparable to other lines (Fig. 2B). We found this interesting because ARR10 transcript is normally present in the wild-type root (Mason et al., 2004; Tajima et al., 2004) and because none of the other type-B ARRs gave a similar hypersensitive phenotype when expressed from the ARR1 promoter. We therefore examined the hypersensitive phenotype of the ARR10 transgenic lines in more detail. Based on a dose response analysis for root growth to cytokinin, the ARR10 lines exhibit hypersensitivity at all cytokinin levels assessed, from 0.001 to 1 µm BA (Fig. 4A). At 1 µm cytokinin, virtually no root growth was observed in the ARR10 lines (Figs. 2B and 4A), suggesting an almost complete absence of cell division in the root (Argyros et al., 2008).
Figure 4.
ARR10 confers cytokinin hypersensitivity when expressed in the same context as ARR1. A, The root elongation response of seedlings grown on media containing 0.001, 0.01, 0.1, and 1 μm BA are expressed as a percentage of the root growth of siblings grown on DMSO control media. Root growth was measured from day 4 through day 7. Error bars indicate se. The mean root growth measurements from untreated lines were 21.1 mm (wild type), 22.4 mm (arr1 arr12), 18.6 mm (tARR1 L2), 19.5 mm (tARR1 L5), 19.2 mm (tARR10 L7), and 19.2 mm (tARR10 L8). B, Induction of callus formation and greening. Representative hypocotyl segments treated with 0.2 mg L–1 indole-3-butyric acid and the indicated concentrations of trans-zeatin are shown after growth for 3 weeks under constant light. C, Relative ARR15 transcript levels in RNA isolated from roots of 14-d-old seedlings treated for 2 h with 10 µm BA or a DMSO control. β-tubulin-3 (At5g62700) was used as an internal control. Transgenic lines tARR1 L5 and tARR10 L7 were examined. D, Protein levels and degradation of ARR1 and ARR10 proteins in Arabidopsis protoplasts. Equal quantities of ARR1 and ARR10 plasmids were transfected into protoplasts. The transfected cells were treated with cycloheximide to inhibit protein biosynthesis, in the absence (–) or presence (+) of trans-zeatin, for the indicated times. ARR1 and ARR10 protein levels were determined by immunoblot analysis with an anti-HA antibody. α-Tubulin protein was immunologically detected as the loading control. Wt, Wild type; CHX, cycloheximide.
Cytokinin also plays a key role in shoot regeneration; therefore, we examined how well ARR10 functionally substitutes for ARR1 in shoot induction assays. As shown in Figure 4B, wild-type tissue demonstrates increased cell division and greening in response to cytokinin treatment, and this response is substantially decreased in the arr1 arr12 mutant, similar to previous results (Mason et al., 2005). Transgenic expression of ARR1 or ARR12 in the arr1 arr12 background rescues the shoot induction phenotype to a similar level as that found in the wild type (Fig. 4B). The proARR1:myc:ARR10 transgene not only rescues the arr1 arr12 mutant, but also results in hypersensitivity to cytokinin in this assay based on two criteria. First, the proARR1:myc:ARR10 transgenic lines exhibit more substantial greening and shoot development than is evident in any of the other lines. Second, the increased greening and shoot induction from callus occurs at lower levels of cytokinin than with the other lines (Fig. 4B).
In tandem with the physiological response phenotypes, we examined molecular responses of the proARR1:myc:ARR10 transgenic line to determine how gene regulation correlates with functional complementation of the arr1 arr12 mutant. Consistent with the physiological responses to cytokinin (Fig. 4, A and B), ARR10 expression resulted in a dramatically increased ARR15 transcript level following cytokinin treatment (Fig. 4C). However, the basal levels of ARR15 in the ARR10 transgenic line were similar to that found in wild-type roots and the arr1 arr12 line functionally complemented with ARR1. This result suggests that the cytokinin hypersensitivity found in the ARR10 line is due to an enhanced ability to mediate cytokinin-regulated gene expression.
Although the transcript levels were comparable among the lines analyzed (Fig. 2B), protein levels tended to be higher for ARR10 compared with the other detectable type-B ARRs (Fig. 2C). In particular, we consistently observed higher protein levels for ARR10 compared with ARR1, even though both were driven from the ARR1 promoter. This raised the question as to whether ARR10 protein is more stable than ARR1 protein, which could potentially account for its increased efficacy in functional complementation experiments. Low protein levels of ARR1 in the transgenic lines precluded a direct examination of protein stability in these lines. Therefore, to test this hypothesis, we transiently transfected Arabidopsis protoplasts with epitope-tagged versions of ARR1 and ARR10 and then examined their protein stability following treatment with the protein biosynthesis inhibitor cycloheximide (Fig. 4D). We observed that ARR10 was degraded more slowly than ARR1 following cycloheximide treatment (Fig. 4D). Treatment with cytokinin did not substantially alter the rate of ARR1 and ARR10 degradation. These results indicate that there is an intrinsic difference in the native protein stability of ARR1 compared with ARR10, consistent with the findings of Kim et al. (2012), which may account for the hypersensitivity observed in the proARR1:myc:ARR10 transgenic lines.
Analysis of Subfamily 2 and 3 Family Members Indicates That ARR21 Can Functionally Complement the arr1 arr12 Mutant
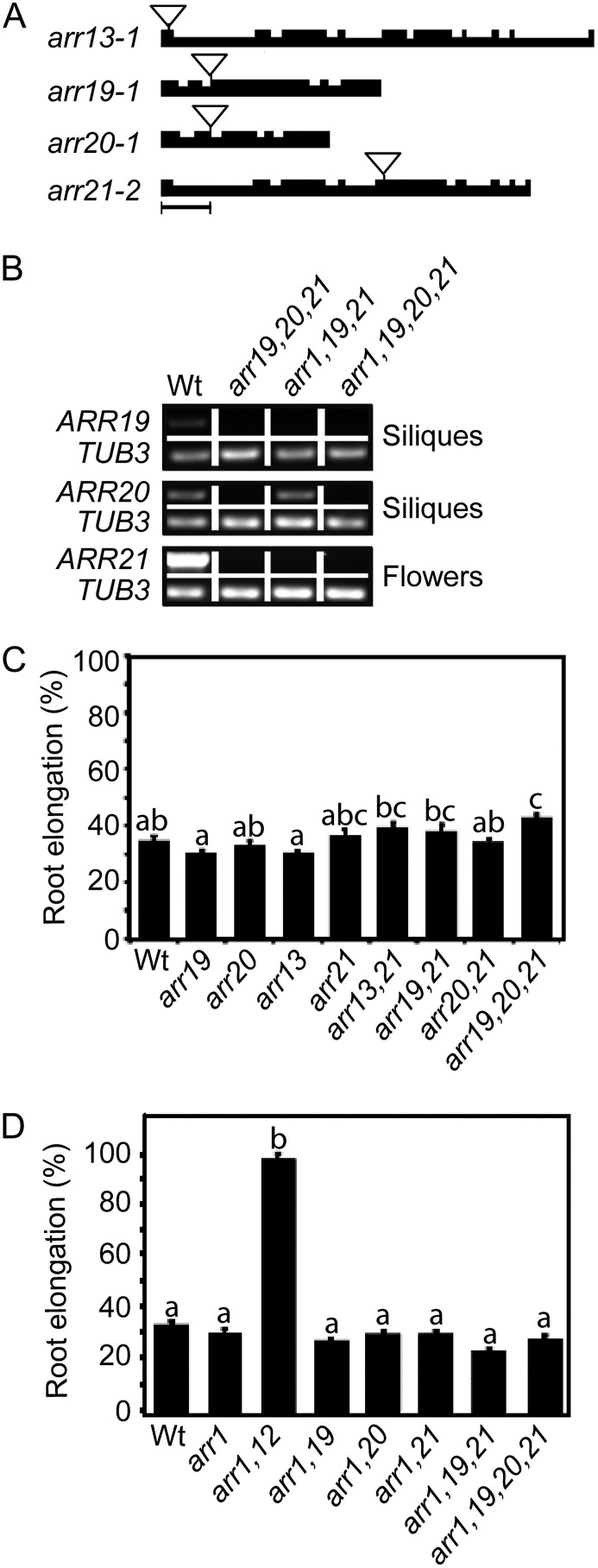
We previously examined transfer DNA (T-DNA) insertion mutants in members of subfamily 1 and determined roles for ARR1, ARR2, ARR10, ARR11, and ARR12 in several cytokinin-mediated responses, including root growth regulation (Mason et al., 2005; Argyros et al., 2008). The subfamily 2 and 3 type-B ARRs exhibit more limited expression than members of subfamily 1 (Mason et al., 2004), but this does not necessarily imply a lack of substantive contribution to plant growth and development. To determine the role of subfamily 2 and 3 members in plant growth, we isolated T-DNA insertion mutants in subfamily 2 members ARR13 and ARR21 and subfamily 3 members ARR19 and ARR20 (Fig. 5A). Because subfamily 2 and 3 members are expressed primarily in the reproductive parts of the plant (Mason et al., 2004), we isolated RNA from these tissues to determine expression levels in the mutants. From RT-PCR analysis of arr19-1, arr20-1, and arr21-2, we determined these mutant lines do not accumulate detectable levels of full-length transcript (Fig. 5B). Lack of transcript is not shown for arr13-1 because transcript was undetectable in wild-type tissues, as previously reported (Mason et al., 2004). As described below, we were able to express detectable ARR13 transcript from the ARR1 promoter, and we were able to use these lines to confirm efficacy of the ARR13 oligonucleotides for expression analysis (Supplemental Fig. S2). Examination of the single and higher order mutants of subfamilies 2 and 3 revealed no pronounced effects on cytokinin sensitivity in root growth assays (Fig. 5C; Supplemental Fig. S3) or hypocotyl elongation assays (Supplemental Fig. S3). We did not observe any obvious effects on flower development, silique development, or seed size in the mutants.
Figure 5.
T-DNA insertion mutants of subfamilies 2 and 3 have minimal effect on cytokinin responses. A, Schematic of the T-DNA insertions in type-B ARR subfamily 2 and 3 genes, arr13-1, arr19-1, arr20-1, and arr21-2. Bar = 500 bp. B, RT-PCR showing lack of ARR19, ARR20, and ARR21 transcript levels in the siliques and flowers (as indicated) of arr19-1, arr20-1, and arr21-2 mutant lines. ARR13 (not shown) was undetectable in the wild type, even after 40 amplification cycles as previously reported (Mason et al., 2004). β-tubulin3 (At5g62700) was used as a loading control. C, Effect of subfamily 2 and 3 mutants on cytokinin sensitivity of the root. The root growth of seedlings grown on media containing 0.1 μm BA is expressed as a percentage of the growth of siblings grown on DMSO control media. Root growth was measured from day 4 through day 7. Error bars indicate se. D, Subfamily 2 and 3 mutations exhibit no additive effects on the cytokinin sensitivity of the root when combined with arr1-3. Lines were analyzed for significant differences in their responsiveness to cytokinin based on Tukey’s multiple range test among the means on the ANOVA (P < 0.05). Lines designated with the same letter exhibit no significant difference.
As an alternative approach to characterize the subfamily 2 and 3 mutations, we generated higher order mutants involving the subfamily 1 mutation arr1-3. These higher order mutants were made with arr1-3, as it represents a sensitized background for mutant analysis, exhibiting a similar cytokinin sensitivity to the wild type as a single mutant (Fig. 5D) but cytokinin hyposensitivity in combination with other subfamily 1 mutations, such as in the arr1 arr12 double mutant (Fig. 5D; Mason et al., 2005; Argyros et al., 2008). However, no additive effects were observed when the loss-of-function alleles of subfamily 2 or 3 members were combined with arr1-3 in a root growth assay (Fig. 5D; Supplemental Fig. S3). A small additive effect was found when arr19 or arr21 were combined with arr1 in a hypocotyl elongation assay, but this was not present in the higher order mutant combinations with arr1 (Supplemental Fig. S4). Thus, no definitive role for subfamily 2 or 3 members in cytokinin-regulated growth was revealed based on several assays.
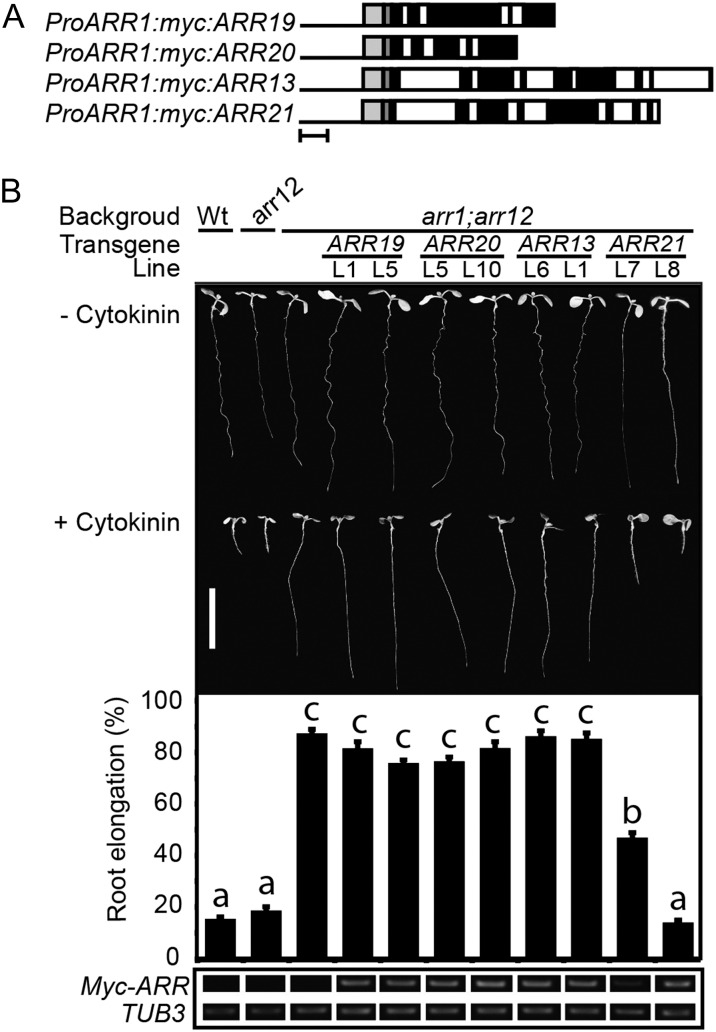
The lack of an effect of the subfamily 2 and 3 mutations is likely due in part to their limited expression pattern in the plant (Mason et al., 2004), raising the question as to whether they could play a more prominent role in cytokinin signaling if broadly expressed. We therefore employed the same approach we took with the subfamily 1 type-B ARRs to determine which could functionally substitute for ARR1. Members of subfamilies 2 and 3 were expressed from the ARR1 promoter and multiple independent transgenic lines assayed for their ability to functionally complement the arr1 arr12 mutant phenotypes. We found that ARR19 (0/4 lines) and ARR20 (0/10 lines) of subfamily 2 and ARR13 (0/8 lines) of subfamily 3 were unable to functionally substitute for ARR1 in root growth assays. Surprisingly, ARR21 (5/8 lines) could functionally substitute for ARR1 in the root growth assay, giving either partial or complete restoration of cytokinin sensitivity. Data for a subset of these lines is shown in Figure 6B, with a line capable of rescue being included if any such was observed. A similar trend was also observed in the ability of the subfamily 2 and 3 ARRs to rescue the enlarged seed size phenotype found in arr1 arr12, with only a transgenic ARR21 line (line 8) clearly demonstrating rescue (Supplemental Fig. S5). By contrast, all the subfamily 2 and 3 members demonstrated little or no ability to functionally substitute for ARR1 in a hypocotyl elongation assay (Supplemental Fig. S5). The inability of ARR19, ARR20, and ARR13 to rescue the arr1 arr12 mutant is not due to poor transgene expression, as they were expressed at comparable levels to the ARR21 transgene that rescued the mutant (Fig. 6B). From these data, we conclude that ARR21 can function in cytokinin signal transduction in a similar capacity to the subfamily 1 members ARR1, ARR2, ARR10, and ARR12 and that the lack of mutant phenotypes in arr21 is likely due to the restricted expression of ARR21 and its functional redundancy with other type-B ARRs.
Figure 6.
Analysis of subfamily 2 and 3 family members indicates that ARR21 can functionally complement the root growth phenotype of the arr1 arr12 mutant. A, Schematic of the subfamily 2 and 3 constructs used in this study. Bar = 500 nucleotides. Black line indicates ARR1 promoter, light-gray box indicates ARR1 5′-UTR, dark-gray box indicates myc sequence, black boxes indicate exons, and white boxes indicate introns. B, The top portion shows representative 7-d-old seedlings grown in presence or absence of 1 µm BA. Bar = 1 cm. The middle portion shows the root elongation response of seedlings grown on media containing 1 μm BA expressed as a percentage of the root growth of siblings grown on DMSO control media. Error bars represent se. The bottom portion shows transcript levels of the ARR transgenes in the roots of 7-d-old seedlings, based on RT-PCR from the sequence encoding the Myc epitope tag. β-tubulin3 (At5g62700) was used as a loading control. Lines were analyzed for significant differences in their responsiveness to cytokinin based on Tukey’s multiple range test among the means on the ANOVA (P < 0.05). Lines designated with the same letter exhibit no significant difference in their responsiveness to cytokinin.
DISCUSSION
Previous work to functionally characterize the type-B ARRs has primarily employed a mutant-based approach to assess their contributions to plant growth and development (Mason et al., 2005; Yokoyama et al., 2007; Argyros et al., 2008; Ishida et al., 2008). However, such an approach is limited because a lack of mutant phenotype may arise due to the genes having low levels of expression, restricted expression patterns, and/or uncharacterized roles in plant development. The type-B ARRs for which functions have been determined are also those that exhibit the broadest expression pattern in Arabidopsis (Mason et al., 2004; Tajima et al., 2004). Subfamily 1 members ARR1, ARR10, and ARR12, in particular, play the most predominant role in regulation of the cytokinin response, consistent with their having a broad expression pattern and also being among the most highly expressed type-B ARRs (Argyros et al., 2008; Ishida et al., 2008). Previous work has also indicated that differences in the temporal expression of ARR1 and ARR12 affect their contribution to the regulation of cell division in the root meristem. Based on the analysis of GUS fusions, expression of ARR1 was noted to increase following germination, while expression of ARR12 remained constant, and this difference correlated with the effects of arr1 and arr12 mutants on root meristem cell number (Dello Ioio et al., 2008b; Moubayidin et al., 2010). We have now extended this analysis by taking a quantitative approach to assess temporal changes in expression at the root tip for the five most abundant type-B ARRs and then correlating these data with the effects of the single mutants. Our analysis confirms the prior data on ARR1 and ARR12 and also indicates that ARR10, which like ARR1 exhibits a temporal increase in expression following germination, behaves similarly to ARR1 in control of meristem cell division based on mutant analysis. ARR2 and ARR11 play less pronounced roles, consistent with their lower levels of expression in the root. The overlapping role in the control of cell division is likely to be mediated through the common mechanism of transcriptional control of SHORT HYPOCOTYL2 (SHY2), a suppressor of the auxin response and a transcriptional target of ARR1 and ARR12 (Dello Ioio et al., 2008b; Moubayidin et al., 2010). Thus, overall, the contribution of type-B ARRs to the cytokinin response closely correlates with their pattern and levels of expression.
As a means to assess functional similarity within the same developmental context, we expressed all 11 type-B ARRs from the ARR1 promoter and determined which could rescue the cytokinin insensitivity phenotype observed in the arr1 arr12 mutant. Results from our studies demonstrate substantial similarity in function among subfamily 1 members ARR1, ARR2, ARR10, and ARR12 and the subfamily 2 member ARR21, all of which can rescue multiple defects found in the arr1 arr12 mutant. The finding that ARR2 and ARR21 exhibit this level of functional similarity is significant, as these type-B ARRs do not display strong mutant phenotypes; thus, their level of contribution to cytokinin signaling is apparently restricted due to their reduced expression profile (Mason et al., 2004; Tajima et al., 2004). Furthermore, a subfamily 1 type-B response regulator from rice (Oryza sativa), one phylogenetically related to ARR10 and ARR12, also restores cytokinin sensitivity to arr1 arr12, indicating a conserved function for some members of this group between monocots and dicots (Tsai et al., 2012).
The functional similarity of subfamily 1 members ARR1, ARR2, ARR10, and ARR12 is likely related to their ability to regulate a similar set of transcriptional targets, as in vitro studies indicate that the DNA-binding domains of ARR1, ARR2, and ARR10 all bind to a core AGATT sequence (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003; Taniguchi et al., 2007). However, whereas ARR1, ARR2, ARR10, and ARR12 are closely related based on phylogenetic analysis, ARR21 is substantially diverged, raising the question as to why it complements the arr1 arr12 mutant but not other more closely related type-B ARRs. The complementation we observe for ARR21 is consistent with previous ectopic studies in which activated versions, lacking their inhibitory receiver domains, of both ARR1 and ARR21 resulted in seedlings that displayed severe developmental abnormalities, such as disordered cell division, along with induction of known cytokinin primary-response genes (Sakai et al., 2001; Tajima et al., 2004; Kiba et al., 2005). Sequence analysis of the DNA-binding domains does not suggest any specific residues that correlate with the ability of type-B ARRs to rescue arr1 arr12 (Tsai et al., 2012). There is, however, a high degree of variation outside of the conserved receiver and DNA-binding domains; thus, more complex interactions not readily identifiable based on sequence homology may play a role in the ability of ARR21 to rescue the mutant phenotype. The finding that ARR21, a diverged member of the type-B ARR family, can complement arr1 arr12 suggests that all members of the family may function as transcription factors, even though this has not been functionally demonstrated for all members.
Six of the type-B ARRs (ARR11, ARR14, and ARR18 of subfamily 1; ARR13 of subfamily 2; and ARR19 and ARR20 of subfamily 3) were unable to rescue the arr1 arr12 mutant, pointing to functional difference(s) among the 11 type-B ARRs of Arabidopsis. These similarities and differences likely relate in part to the ability of the type-B ARRs to transcriptionally regulate an overlapping set of primary-response genes, based on the known function of type-B ARRs in transcriptional regulation (Sakai et al., 2000, 2001; Imamura et al., 2001, 2003; Lohrmann et al., 2001; Hosoda et al., 2002; Mason et al., 2004, 2005; Rashotte et al., 2006; Liang et al., 2012; Tsai et al., 2012). DNA-binding studies indicate that ARR11, unlike ARR1, ARR2, and ARR10, does not bind to the core AGATT sequence (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003; Taniguchi et al., 2007). Similarly, in yeast (Saccharomyces cerevisiae) one-hybrid assays, ARR2 but not ARR11 associates with an anther/pollen-specific promoter fragment (Lohrmann et al., 2001). These data are consistent with our finding that ARR11 cannot functionally substitute for ARR1 or ARR12 and suggest that this arises in part from differences in their target specificity. The inability of ARR18 to complement arr1 arr12 also likely arises in part due to differences from ARR1 in terms of target affinity or specificity based on our transcriptional analysis. Whereas transgenic expression of ARR1 in arr1 arr12 facilitated cytokinin-mediated induction of the primary-response genes ARR5 and ARR15 and cytokinin-mediated suppression of HKT1, transgenic expression of ARR18 only facilitated the suppression of HKT1. In addition, although we observed that ARR18 functioned as a transcription factor in a transient protoplast expression system, it differed from ARR1 and ARR12 in that it did not activate the ARR6 reporter in a cytokinin-dependent fashion. Differences in the ability of the type-B ARRs to transcriptionally regulate targets need not only arise from individual differences in target affinity or specificity, but could also arise from differences in type-B ARR protein stability (Kim et al., 2012) or their interactions with upstream regulators such as the ARABIDOPSIS HIS-CONTAINING PHOSPHOTRANSFER PROTEINS and/or transcriptional coregulators (Dortay et al., 2006; Kim et al., 2006). For example, ARR2 appears to be specifically activated through an AHK3-dependent phosphorelay in the regulation of leaf senescence (Kim et al., 2006). A potential lack of the relevant regulators would preclude the activation of the type-B ARRs.
The inability of ARR11, ARR14, ARR18, ARR19, and ARR20 to complement the arr1 arr12 mutant phenotype is not what one would necessarily predict based on previous characterization using transient protoplast assays and overexpression analysis in wild-type plants. In protoplasts, ARR14 and ARR20 stimulated a luciferase reporter driven by a concatamerized type-B binding site (TWO COMPONENT OUTPUT SENSOR::LUC) in a cytokinin-dependent manner, whereas ARR19 stimulated expression of TWO COMPONENT OUTPUT SENSOR::LUC in a cytokinin-independent manner, as we saw with ARR18 using the ARR6::LUC reporter (Müller and Sheen, 2008; Zürcher et al., 2013). In a separate protoplast assay, ARR18 induced an ARR5:LUC construct by about 50% in the presence of cytokinin (Veerabagu et al., 2012). When ectopically overexpressed in transgenic plants, ARR18 increased the cytokinin sensitivity, and activated versions of ARR11, ARR18, and ARR19 induced a cytokinin-like response (Liang et al., 2012; Veerabagu et al., 2012). Two possibilities, not mutually exclusive, can explain the differences observed between these studies and ours. First, the type-B ARRs were overexpressed in these previous studies, rather than being expressed from the ARR1 promoter, high levels of the type-B ARRs potentially allowing for cross talk with the cytokinin-signaling pathway and/or overcoming a reduced affinity for the target DNA sites of ARR1. Second, these previous studies were performed in a wild-type background, as opposed to the arr1 arr12 background, raising the possibility that their function is dependent in part on genes regulated through action of ARR1 and/or ARR12. Alternatively, because ARR18 multimerizes (Veerabagu et al., 2012), a physical association with ARR1 and/or ARR12 may allow for indirect transcriptional regulation. Characterization of two-component signaling in plants is likely to be particularly susceptible to artifacts from overexpression based on the known potential for promiscuous interactions in two-component systems (Skerker et al., 2008; Bell et al., 2010; Schaller et al., 2011). This last point is demonstrated by the ability of Arabidopsis cytokinin receptors and response regulators to function within a bacterial two-component system when transgenically expressed in Escherichia coli (Imamura et al., 1998; Yamada et al., 2001).
The observed differences in the ability of type-B ARRs to regulate gene expression and restore physiological responses in planta raises the question as to the function of the other type-B ARRs. It is likely that some also participate in cytokinin signaling, but (1) due to lower affinity for their targets, primarily play a role at high cytokinin levels; (2) due to higher rates of turnover, have a proportionately reduced contribution; (3) require additional coregulators to mediate their effects on transcription; and/or (4) regulate expression of different target genes than those regulated by the type-B ARRs implicated in cytokinin signaling. For example, whereas ARR18 did not functionally complement the arr1 arr12 mutant, we did find evidence that ARR18 could regulate a subset of cytokinin-dependent genes in planta. It may also be that some type-B ARRs do not primarily function in cytokinin signaling but regulate the transcriptional response of other plant His kinases, such as AHK1, implicated in the osmotic response (Tran et al., 2007) or CYTOKININ-INDEPENDENT1 implicated in embryogenesis (Pischke et al., 2002; Hejátko et al., 2003). A better understanding of the targets of these type-B ARRs will likely provide key information on how they participate in two-component signaling pathways in plants.
Interestingly, and unlike the case with any of the other type-B ARRs, we found that expression of ARR10 in the context of ARR1 results in hypersensitivity to cytokinin. This level of hypersensitivity is greater than that reported from overexpression of ARR1, which showed slightly increased sensitivity to low concentrations of cytokinin but was comparable to the wild type at higher concentrations (Sakai et al., 2001). The cytokinin hypersensitivity in the lines expressing ARR10 likely arises from the change in the zone of ARR10 expression combined with the enhanced stability of the ARR10 protein. Based on GUS fusions and transcriptional profiling of the primary root tip, ARR1 is expressed at similar levels throughout the stele, endodermis, cortex, and epidermis, but ARR10 is expressed at higher levels in the epidermis than in the other tissues (Birnbaum et al., 2003; Mason et al., 2004; Argyros et al., 2008). Thus, when driven from the ARR1 promoter, the level of ARR10 within the internal tissues of the root would increase dramatically. In the transgenic plants, ARR10 protein accumulates to higher levels than ARR1 even though transcript levels are similar, apparently due to their differing rates of protein degradation. A higher level of ARR10 protein with a slower rate of degradation would allow for a greater number of ARR10 proteins to become activated in response to cytokinin and consequently increase the level and duration of the transcriptional response to cytokinin, as previous research has demonstrated that increasing the expression levels of type-B ARRs can enhance the cytokinin sensitivity of transgenic lines (Sakai et al., 2001; Liang et al., 2012). Other mechanisms may also contribute to the hypersensitivity conferred by ARR10, but the posttranscriptional difference in degradation rates between ARR10 and ARR1 alone is predicted to result in an enhanced efficacy for mediating the cytokinin signal.
The cytokinin hypersensitivity conferred by ARR10 has potential agronomic benefits, in particular the ability to increase cytokinin sensitivity in tissue culture. There is considerable variability in regenerative potential observed among plant species, even among different lines of Arabidopsis and rice (Abe and Futsuhara, 1986; Candela et al., 2001; Khalequzzaman et al., 2005), and this can pose a significant problem for transformation of crop plants (Birch, 1997). It was previously found that loss of four or more type-A ARRs, which serve to negatively regulate cytokinin signaling, resulted in increased regenerative potential for tissue culture (To et al., 2004). Here, we find that a change in the expression pattern for a single type-B ARR, ARR10, has a profound effect on regenerative capacity, suggesting that either ARR10 itself or orthologs from other plant species may be used to circumvent the recalcitrance of some crop species to tissue culture techniques.
MATERIALS AND METHODS
Plant Growth Conditions and Growth Assays
Wild-type and mutant lines of Arabidopsis (Arabidopsis thaliana) are all in the Columbia ecotype and were grown as previously described (Argyros et al., 2008). Root growth, hypocotyl elongation, shoot induction, and seed size assays were performed as previously described (Mason et al., 2005; Argyros et al., 2008).
Constructs and Generation of Transgenic Lines
To express the type-B ARRs from the ARR1 promoter in planta, we constructed the binary destination vector, pEARLEY-pARR1:myc-Gateway cassette (GW), which contained an ARR1 promoter, a c-myc epitope tag, and a Gateway cloning site. To construct this vector, a 1.2-kb region corresponding to the ARR1 promoter, 5′-untranslated region (UTR), and ATG start codon was PCR amplified from genomic DNA using the primers 5′-CTAATCATAGTTACACACGACTTG-3′ and 5′-CATACCTCTCTCTATGTAGCTCG-3′ and ligated into the pCR8 entry vector (Invitrogen K2520–02) according to manufacturer’s instructions. We then moved the ARR1 promoter from the entry vector into the destination vector pEarleyGate303 (Earley et al., 2006) by Gateway technology, thus generating the pEARLY-pARR1-myc intermediate. The SpeI/BglII fragment containing a Gateway cloning site was isolated from pEarleyGate203 (Earley et al., 2006) and cloned into the analogous restriction sites of pEARLY-pARR1-myc to generate pEARLY-pARR1:myc-GW.
For expression in plants, the 11 type-B ARR sequences were amplified from Arabidopsis Columbia genomic DNA using oligonucleotides (Supplemental Table S1) that amplified from the translational start codon and included the stop codon, ligated into the pCR8 entry vector, and moved into pEARLY-pARR1:myc-GW according to the manufacturer (Invitrogen). The pEARLEY-pARR1:myc-ARR constructs were confirmed by sequencing and introduced into the arr1-3 arr12-1 double mutant by the floral dipping method (Bent and Clough, 1998) using the Agrobacterium tumefaciens strain GV1301. For the protoplast transactivation assay constructs, the 35S promoter-GW-octapine synthase fragment from pEarleygate100 and the 35S promoter-myc tag-GW fragment from pEarleygate203 (Earley et al., 2006) were amplified (primers 5′-TAGGTACCGAATTCCAATCCCACAAAAATCTG-3′ and 5′-TAAAGCTTGGTCCTGCTGAGCCTCGA-3′) and cloned into the pBluescript II KS+ (pBS) vector (Agilent Technologies) through KpnI and HindIII restriction sites to generate the Gateway compatible overexpression vectors pBS-35S-GW and pBS-35S-myc-GW. The genomic fragments of ARR1 (primers 5′-ATGATGAATCCGAGTCACGGAA-3′and 5′-AACCTGCTTAAGAAGTGCGCTC-3′), ARR12 (primers 5′-CACCTCTGATCCGAACAATGGGAAAGG-3′ and 5′-TCATATGCATGTTCTGAGTGAACTAAAC-3′), and ARR18 (primers 5′-ATGAGGGTTCTTGCTGTGGAT-3′ and 5′-CTAAGGTGGAGGAAATGAATCAAAGC-3′) were amplified and cloned into the pCR8/GW/TOPO vector (Invitrogen) to generate the entry clones and then recombined into pBS-35S-GW and pBS-35S-myc-GW for protoplast luciferase assays.
For protein stability assays in protoplasts, complementary DNA sequences for ARR1 (primers 5′-GGATCCATGATGAATCCGAGTCACGGAAGA-3′ and 5′-AGGCCTAACCTGCTTAAGAAGTGCGCTC-3′) and ARR12 (primers 5′-CCATGGCTATGGAGCAAGAAATTGAAGTC-3′ and 5′-AGGCCTAGCTGACAAAGAAAAGGGAAAATG-3′) were fused to the hemagglutinin (HA) epitope coding sequence and expressed from the 35SC4PPDK promoter as described (Sheen, 1996).
T-DNA Insertion Lines
The subfamily-1 mutant alleles have been previously described (Mason et al., 2005; Argyros et al., 2008), except for arr2-5 (GABI_269G01). The arr13-1, arr19-1, and arr20-1 T-DNA mutant alleles were initially identified by PCR-based screening approaches with a T-DNA insertion population as described (Alonso et al., 2003; Mason et al., 2005), with arr13-1 (SALK_042719) and arr20-1 (SALK_009734) both available now from the Salk Collection. The mutant allele arr21-2 (SALK_005772) was obtained from the Salk Collection. Sequence analysis of arr2-5 identified the T-DNA junction with ARR2 as (tacaattgaatatatcctg)tcgttgaatactcatTGCGAATCTTCGAGTTCTTGT, with uppercase letters indicating the ARR2 sequence and parentheses indicating the T-DNA left border sequence, placing the insertion site within the first exon. Sequence analysis of arr13-1 identified the T-DNA junction with ARR13 as GTTGTGGACGATAATCGTGTT(gtaaacaaattgacgcttaa), placing the insertion site within the first exon. Sequence analysis of arr19-1 identified the T-DNA junction as CACAATCTATTTCATATTTGTGa(tgtaaacaaattgacgct), placing the insertion site within the second intron. Sequence analysis of arr20-1 identified the T-DNA junction as ACCCGTAGTAAGTAAGTATATtggacgt(tattgtggtgtaaacaaattg), placing the insertion site within the second intron. Sequence analysis of arr21-2 identified the T-DNA junction as (ttgtctaagcgtcaatttgt)TCACATTAAGGAGCCGTACTT, placing the insertion site within the fifth exon.
RNA Expression Analysis
Total RNA was isolated and first-strand complementary DNA synthesis performed as previously described (Argyros et al., 2008). RT-PCR was used to confirm lack of RNA expression in the T-DNA insertion lines, and the primer sequences used for this analysis can be found in Supplemental Table S2. Semiquantitative RT-PCR was used to confirm and compare expression of the transgenes driven by the ARR1 promoter. For this purpose, we used a primer against the 5′-untranslated region of ARR1 (5′-GAGATTCACTTCTATCTCCAACAATTTCG-3′) and against the c-myc epitope tag (5′-CAAACTTGTGATCAGATCTTCTTCAGAG-3′). In addition, the presence of full-length transcript was verified for the transgenic lines using gene-specific primer pairs (data not shown). Quantitative real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa Bio, RR041A) according to the manufacturer, as previously described using primer pairs specific for the genes of interest (Supplemental Table S3). β-TUBULIN3 (At5g62700) was used as a loading and normalization control for RT-PCR and quantitative real-time PCR with primers 5′-TGGTGGAGCCTTACAACGCTACTT-3′ and 5′-TTCACAGCAAGCTTACGGAGGTCA-3′.
Transactivation and Protein Stability Assays in Arabidopsis Protoplasts
Arabidopsis protoplasts were isolated and transfected as described (Hwang and Sheen, 2001; Yoo et al., 2007). For transactivation assays, the ARR6-LUC reporter gene was transfected alone or cotransfected with ARR1, ARR12, or ARR18 effectors into protoplasts isolated from wild-type plants. Transfected protoplasts were incubated with or without 100 nm trans-zeatin for 3 h under dim light (5 µE m–2 s–1). The UBIQUITIN10-GUS construct was used as an internal control. For protein stability assays, transfected protoplasts were incubated for 4 h to allow for protein expression and then treated with 100 μm of cycloheximide and 1 µm of trans-zeatin for indicated times.
Protein Isolation and Immunoblot Analysis
Seedling samples were ground in liquid nitrogen and the powder resuspended in isolation buffer containing 50 mm TRIS (pH 7.5), 0.1% (v/v) Nonidet P-40, 10 mm EDTA, 150 mm NaCl, and protease inhibitors (Sigma-Aldrich, P9599). Protoplast samples were frozen, resuspended in isolation buffer, and vortexed. Samples were centrifuged at 16,000g for 15 min and the supernatant retained for further analysis. Protein concentration was determined by use of the bicinchoninic acid reagent (Pierce) according to the manufacturer after first adding 0.2% (w/v) SDS to the samples and with bovine serum albumin as a standard. Samples were heated above 65°C in gel-loading buffer, and SDS-PAGE and immunoblotting was performed as previously described (Gao et al., 2008). HA-tagged proteins were detected by using a peroxidase-conjugated anti-HA antibody (Roche Applied Science). Myc-tagged proteins were detected with a monoclonal anti-Myc antibody conjugated to horseradish peroxidase (monoclonal 9E-10; Santa Cruz Biotechnology). Monoclonal antibodies against Hsc70 protein (StressGen) and α-tubulin (Sigma-Aldrich), coupled with goat anti-mouse IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology), were used for detection of these protein-loading controls.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ARR1 (At3g16857), ARR2 (At4g16110), ARR10 (At4g31920), ARR11 (At1g67710), ARR12 (At2g25180), ARR13 (At2g27070), ARR14 (At2g01760), ARR18 (At5g58080), ARR19 (At1g49190), ARR20 (At3g62670), ARR21 (At5g07210), ARR5 (At3g48100), ARR15 (At1g74890), and HKT1 (At4g10310).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A subset of subfamily 1 ARRs can functionally complement the arr1 arr12 mutant based on cytokinin-modulated hypocotyl elongation and seed size.
Supplemental Figure S2. RT-PCR confirmation of transgene expression in transgenic lines of arr1 arr12.
Supplemental Figure S3. T-DNA insertion mutants of subfamilies 2 and 3 have minimal effect on cytokinin response based on a root growth dose response assay.
Supplemental Figure S4. T-DNA insertion mutants of subfamilies 2 and 3 have minimal effect on cytokinin responses in hypocotyl elongation assays.
Supplemental Figure S5. Ability of subfamily 2 and 3 family members to functionally complement the arr1 arr12 mutant based on cytokinin-modulated hypocotyl elongation and seed size.
Supplemental Table S1 Oligonucleotides used for cloning type-B ARRs.
Supplemental Table S2. Oligonucleotides used to examine expression of subfamily 2 and 3 T-DNA insertion lines.
Supplemental Table S3. Oligonucleotides used for quantitative RT-PCR.
Glossary
- Ct
threshold cycle
- RT
reverse transcription
- BA
benzyladenine
- T-DNA
transfer DNA
- HA
hemagglutinin
- DMSO
dimethyl sulfoxide
- GW
Gateway cassette
- UTR
untranslated region
References
- Abe T, Futsuhara Y. (1986) Genotype variability for callus formation and plant regeneration in rice (Oryza sativa L.). Theor Appl Genet 72: 3–10 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CH, Porter SL, Strawson A, Stuart DI, Armitage JP. (2010) Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex. PLoS Biol 8: e1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Clough SJ. (1998) Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In Gelvin SB, Schilperout RA, eds, Plant Molecular Biology Manual, Ed 2 Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–14 [Google Scholar]
- Birch RG. (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Physiol Plant Mol Biol 48: 297–326 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Candela M, Velázquez I, de la Cruz B, Sendino AM, de la Peña A. (2001) Differences in in vitro plant regeneration ability among four Arabidopsis thaliana ecotypes. In Vitro Cell Dev Biol Plant 37: 638–643 [Google Scholar]
- Dello Ioio R, Linhares FS, Sabatini S. (2008a) Emerging role of cytokinin as a regulator of cellular differentiation. Curr Opin Plant Biol 11: 23–27 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. (2008b) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A. (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273: 4631–4644 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Gao Z, Wen C-K, Binder BM, Chen Y-F, Chang J, Chiang Y-H, Kerris RJ, III, Chang C, Schaller GE. (2008) Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem 283: 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ. (2002) Cytokinins. New insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J, Pernisová M, Eneva T, Palme K, Brzobohatý B. (2003) The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol Genet Genomics 269: 443–453 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T. (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. (1998) Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. (2003) In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 122–131 [DOI] [PubMed] [Google Scholar]
- Imamura A, Yoshino Y, Mizuno T. (2001) Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci Biotechnol Biochem 65: 2113–2117 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Khalequzzaman M, Haq N, Hoque ME, Aditya TL. (2005) Regeneration efficiency and genotypic effect of 15 Indica type Bangladeshi rice (Oryza sativa L.) landraces. Plant Tissue Cult 15: 33–42 [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. (2005) Combinatorial microarray analysis revealing arabidopsis genes implicated in cytokinin responses through the His->Asp Phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ryu H, Cho YH, Scacchi E, Sabatini S, Hwang I. (2012) Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR2 attenuates signaling output in two-component circuitry. Plant J 69: 934–945 [DOI] [PubMed] [Google Scholar]
- Liang Y, Wang X, Hong S, Li Y, Zuo J. (2012) Deletion of the initial 45 residues of ARR18 induces cytokinin response in Arabidopsis. J Genet Genomics 39: 37–46 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schäfer E, Kudla J, Harter K. (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE. (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 64: 753–763 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok MC. (1994) Cytokinins and plant development: an overview. In Mok MC, ed, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166 [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke MS, Jones LG, Otsuga D, Fernandez DE, Drews GN, Sussman MR. (2002) An Arabidopsis histidine kinase is essential for megagametogenesis. Proc Natl Acad Sci USA 99: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, Armitage JP. (2011) Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21: R320–R330 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. (2008) Rewiring the specificity of two-component signal transduction systems. Cell 133: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T. (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tran L-SP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, Schaller GE, Kieber JJ. (2012) Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol 158: 1666–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S. (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Veerabagu M, Elgass K, Kirchler T, Huppenberger P, Harter K, Chaban C, Mira-Rodado V. (2012) The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J 72: 721–731 [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB. (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T. (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A, Sakakibara H, Mizuno T. (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48: 84–96 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkeli K, Müller B. (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]