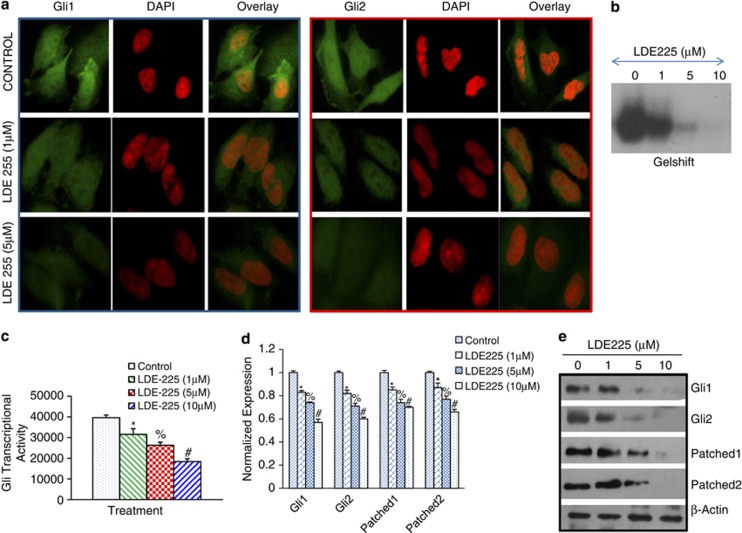
Figure 3.
NVP-LDE-225 downregulates Shh-signaling pathway in CSCs. (a) NVP-LDE-225 inhibits expression of Gli1 and Gli2 in CSCs. The cells were seeded on fibronectin-coated coverslips and treated with NVP-LDE-225 (5 μℳ) for 48 h. Subsequently, cells were fixed with 4% paraformaldehyde, blocked in 5% normal goat serum and stained with Gli1 and Gli2 primary antibodies (1:100) for 16 h at 4 °C and washed with PBS. Afterwards, cells were incubated with fluorescently labeled secondary antibody (1:200) along with DAPI (1 mg/ml) for 1 h at room temperature and cells were mounted and visualized under a fluorescent microscope. (b) Gel shift asssay. Nuclear extracts were prepared and incubated with labeled Gli probe. Gelshift assay was performed as we described in Materials and methods. (c) Luciferase assay, CSCs were transduced with lentiviral particles expressing Gli-dependent luciferase reporter and treated with NVP-LDE-225 (0, 5 and 10 μℳ) for 48 h. Lysates were prepared, and luciferase activity was measured as described in Materials and methods. Normalized luciferase activity is presented as mean±s.d. *, % and #=Significantly different from control (P<0.05). (d) CSCs were treated with NVP-LDE-225 (10 μℳ) for 36 h. At the end of incubation period, RNA was extracted and the expression of Gli1, Gli2, Patched-1 and Patched-2 was measured by qRT–PCR. Data represent the mean±s.d (n=4). *, % and #=Significantly different from respective control (P<0.05). (e) Protein expression of Gli1, Gli2, Patched1 and Patched-2. CSCs were treated with NVP-LDE-225 for 48 h, and the expression of Gli1, Gli2, Patched1 and Patched-2 was determined by the western blot analysis.