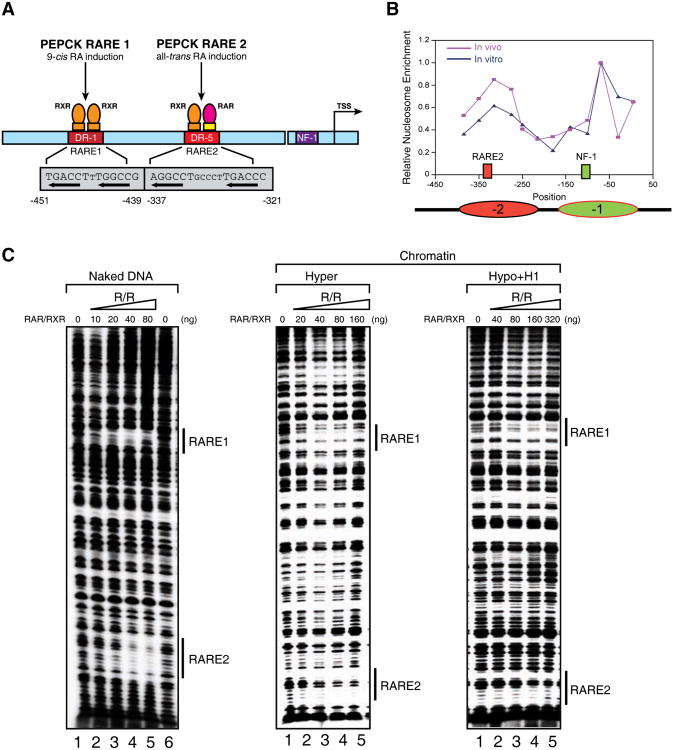
Figure 2.
The effects of chromatin structure on RAR/RXR binding to chromatin templates in vitro.
(A) Diagram of the p(PEPCK-500)G template containing the PEPCK promoter used for in vitro DNase I footprinting and transcription assays. (B) Line graph of nucleosomal positioning at the PEPCK promoter in vivo (red) and in vitro (blue). The nucleosomes covering RARE2 and the NF1 binding site are represented as ovals in red and green, respectively. The mapping of nucleosomal positioning at the PEPCK promoter in vitro and in vivo is described in Experimental Procedures. Nucleosomal positioning in vivo shown here represents the nucleosome structure after tRA treatment (165 min). (C) DNase I footprinting analysis of RAR/RXR bound to the PEPCK promoter on naked DNA (left panel), or chromatin fiber assembled with either hyperacetylated histones (middle panel), or hypoacetylated histones and histone H1 (right panel). The binding sites for RAR/RXR (RARE1 and -2) are indicated by bond bars. See also Figure S2.