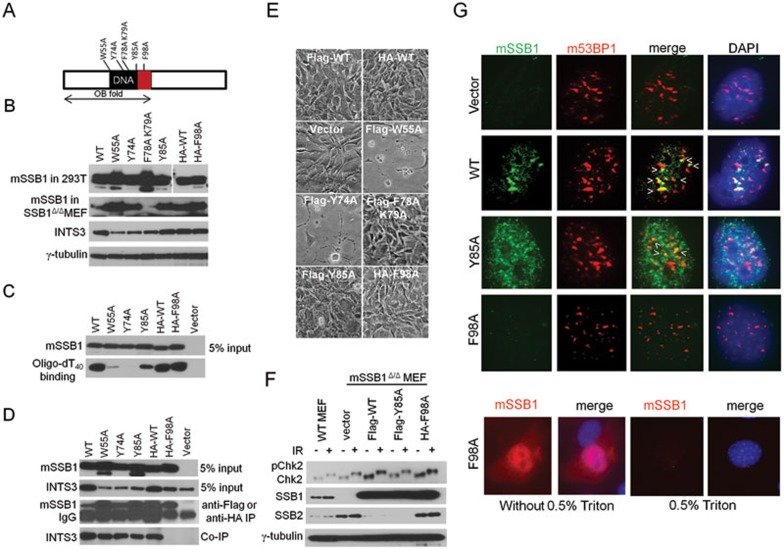
Figure 3.
The mSSB1 OB-fold is required for ssDNA binding and interaction with INTS3. (A) Schematics of conserved amino acid residues postulated to be important for DNA binding and interaction with INTS3 (red box). Residues were mutated individually to alanines. (B) Detection of mSSB1 WT and mutant proteins in 293T cells and mSSB1Δ/Δ MEFs. γ-tubulin served as loading control. (C) ssDNA binding assay. Cell lysates extracted from mSSB1Δ/Δ MEFs expressing mSSB1 WT and mutant proteins were incubated with biotin-oligo-dT40-coated streptavidin-beads. ssDNA-bound mSSB1 proteins were eluted and detected by immunoblotting. (D) Interaction between WT or mSSB1 mutants with endogenous INTS3 in mSSB1Δ/Δ MEFs was assayed by coimmunoprecipitation assays. (E) Morphology of mSSB1Δ/Δ MEFs reconstituted with WT or mutant mSSB1. (F) Immunoblot detection of mSSB1Δ/Δ MEFs reconstituted with the indicated constructs, probed with antibodies to detect phosphorylated Chk2, mSSB1 and mSSB2. γ-tubulin served as loading control. (G) Co-localization of 53BP1 with WT or mSSB1 mutants reconstituted in mSSB1Δ/Δ MEF, following exposure to 5Gy IR. 0.5% Triton treatment of mSSB1Δ/Δ MEFs expressing SSB1F98A reduced nuclear staining.