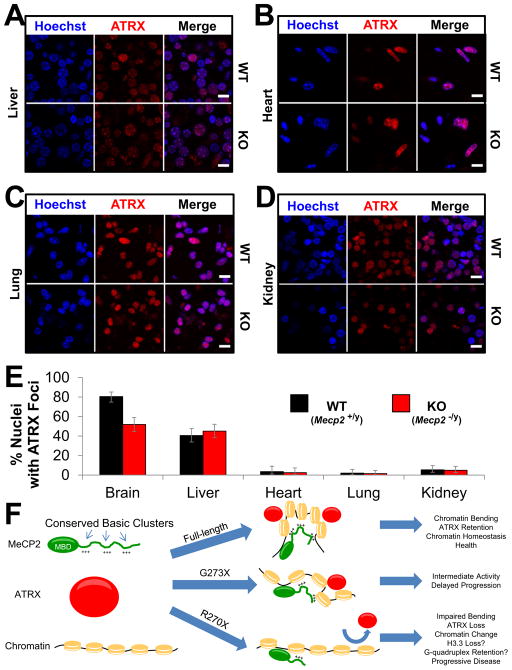
Figure 7.
ATRX mislocalization is specific to the brain
(A–D) Immunofluorescence for ATRX in fresh nuclei prepared from WT and KO mice at 9 weeks of age. Nuclei are counterstained with Hoechst from liver (A), heart (B), lung (C), and kidney (D). Scale bars represent 10 μm. n = 3 mice per genotype. (E) Quantification of the experiment described in (A–D and Figure S7C). The percentage of ATRX positive nuclei with foci localizing to PCH from WT and KO animals is plotted for each tissue. Error bars represent 95% confidence intervals. For brain n = 226 and 183; for liver n = 192 and 195; for heart n = 112 and 121; for lung n = 151 and 195; for kidney n = 184 and 204 nuclei from WT and KO mice respectively. (F) Proposed model for MeCP2 in chromatin homeostasis. MeCP2 contains highly conserved basic clusters (+++) within its disordered C-terminal region. MeCP2 first binds DNA through its MBD, but the presence of multiple DNA binding elements allow MeCP2 to alter chromatin conformation, leading to homeostasis as indicated by the recruitment of ATRX to PCH. When the C-terminus becomes truncated (e.g., G273X) this activity is reduced. Further truncation beyond the AT-Hook 2 domain (e.g., R270X) severely impairs the function of MeCP2, chromatin is no longer maintained in a physiological conformation, and ATRX is lost from PCH. See also Figure S7.