Abstract
Many methods have been proposed for computer-assisted diagnostic classification. Full tensor information and machine learning with 3D maps derived from brain images may help detect subtle differences or classify subjects into different groups. Here we develop a new approach to apply tensor-based morphometry to parametric surface models for diagnostic classification. We use this approach to identify cortical surface features for use in diagnostic classifiers. First, with holomorphic 1-forms, we compute an efficient and accurate conformal mapping from a multiply connected mesh to the so-called slit domain. Next, the surface parameterization approach provides a natural way to register anatomical surfaces across subjects using a constrained harmonic map. To analyze anatomical differences, we then analyze the full Riemannian surface metric tensors, which retain multivariate information on local surface geometry. As the number of voxels in a 3D image is large, sparse learning is a promising method to select a subset of imaging features and to improve classification accuracy. Focusing on vertices with greatest effect sizes, we train a diagnostic classifier using the surface features selected by an ℓ1-norm based sparse learning method. Stability selection is applied to validate the selected feature sets. We tested the algorithm on MRI-derived cortical surfaces from 42 subjects with genetically confirmed Williams syndrome and 40 age-matched controls, multivariate statistics on the local tensors gave greater effect sizes for detecting group differences relative to other TBM-based statistics including analysis of the Jacobian determinant and the largest eigenvalue of the surface metric. Our method also gave reasonable classification results relative to the Jacobian determinant, the pair of eigenvalues of the Jacobian matrix and volume features. This analysis pipeline may boost the power of morphometry studies, and may assist with image-based classification.
Keywords: surface conformal parameterization, surface registration, tensor-based morphometry, sparse learning, stability selection
1. INTRODUCTION
Computer-assisted diagnostic classification is becoming increasingly popular in neuroimaging, especially given the vast number of features available to assist diagnosis in a 3D brain image. Early diagnosis and treatment of degenerative brain diseases, such as Alzheimer’s disease, depends on the ability to identify disease in its earliest stages, when brain changes may be subtle. In addition, there is interest in understanding which brain imaging features are best for diagnostic classification, as well as biomarkers to measure the severity of disease burden. Over the last decade, many methods have been proposed to study the problem of diagnostic classification based on structural magnetic resonance imaging (MRI) (Golland et al., 2001; Yushkevich et al., 2003; Fan et al., 2007; Vemuri et al., 2008; Gutman et al., 2009; Sun et al., 2009a; Xiang et al., 2009; Cuingnet et al., 2010; Cuingnet et al., 2011; Sabuncu and Van Leemput, 2011; Batmanghelich et al., 2012), positron emission tomography (PET) (Chen et al., 2011; López et al., 2011), single photon emitting computer tomography (SPECT) (Fung and Stoeckel, 2007) or a combination of multi-source datasets (Calhoun and Adali, 2009; Chen et al., 2009; Correa et al., 2010; Jack et al., 2010; Kohannim et al., 2010; Yang et al., 2010; Groves et al., 2011; Sui et al., 2011; Yuan et al., 2012). Surface-based modeling is useful in brain imaging to help analyze anatomical shapes, to detect abnormalities in cortical surface folding and thickness, and to statistically combine or compare 3D anatomical models across subjects (Drury et al., 1996; Thompson and Toga, 1996; Fischl et al., 1999; Vaillant et al., 2007; Yeo et al., 2008; Wang et al., 2010c; Wang et al., 2011b). Many surface-based morphometry studies describe structural differences at the group level, i.e., between different diagnostic groups. More recently, morphometric maps have also been used to classify individual subjects into diagnostic groups (Ferrarini et al., 2008; Sun et al., 2009a; Kohannim et al., 2010; Wang et al., 2010b; Costafreda et al., 2011). In one study (Sun et al., 2009a), maps of cortical gray matter density achieved 86.1% accuracy in discriminating psychotic patients from control subjects, in leave-one-out tests. In related work (Ferrarini et al., 2008), the notion of biomarker “nodes” was proposed, i.e. regions on surface meshes that contribute most to diagnostic classification; the authors tested their approach on ventricular surface models from Alzheimer’s disease patients and matched controls. Overall, a set of surface-based morphometric features combined with a machine learning algorithm may offer a promising way to improve the performance of computer-assisted diagnostic systems.
An important question for diagnostic classification based on voxel-based or surface-based morphometric maps is which statistics are best to analyze. Statistics derived from anatomical surface models, such as gray matter thickness maps (Thompson et al., 2003; Thompson et al., 2005), radial distances (distances from the medial core to each surface point) (Styner et al., 2004; Thompson et al., 2004a; Carmichael et al., 2006; Carmichael et al., 2007b; a; Carmichael et al., 2007c; Thompson et al., 2007; Chou et al., 2008; Chou et al., 2009; Morra et al., 2009; Apostolova et al., 2010a; Apostolova et al., 2010b; Morra et al., 2010), spherical harmonic analysis (Styner et al., 2005; Gutman et al., 2009), local area differences (related to the determinant of Jacobian matrix) (Davatzikos et al., 1996; Woods, 2003; Chung et al., 2008), Gaussian random fields (Bansal et al., 2007), Reeb graphs (another way to compute radial distances) (Shi et al., 2009) have all been applied to analyze the shape and geometry of various brain structures. Surface tensor-based morphometry (TBM) (Davatzikos et al., 1996; Thompson et al., 2000a; Woods, 2003; Chung et al., 2008) is an intrinsic surface statistic that examines spatial derivatives of the deformation maps that register brains to common templates, and can help to detect subtle differences in local surface morphometry. In recent studies (Wang et al., 2008b; Wang et al., 2009a; Wang et al., 2010c; Wang et al., 2011b), surface multivariate TBM (mTBM) was found to be more sensitive for detecting group differences than other standard TBM-based statistics. As a result, here we decided to use mTBM statistics as the surface statistics to be included in a diagnostic classifier.
Three-dimensional statistical maps can detect consistent local differences in anatomical surfaces. But, when they are applied to classification, the feature dimension is usually much larger than the number of subjects in the sample being analyzed—the “high dimension/small sample size problem”. When a vast number of variables are measured from a small number of subjects, it is often possible to divide the subjects into groups based on the observed features, but the resulting classification rules may generalize poorly to new observations. To select the most useful features, feature reduction can be beneficial. Feature selection approaches are widely used in machine learning, (e.g. (Stearns, 1976; Guyon et al., 2002; Fan et al., 2005; Kuncheva and Rodríguez, 2010)). Even so, most methods still generate very large numbers of features, making it difficult to state intuitively why features are being used to make biological inferences. To address this, sparse learning methods have been proposed to select the most biologically germane features (Tibshirani, 1996; Friedman et al., 2008). Sparse learning methods enjoy strong theoretical properties (Donoho, 2006; Candès and Wakin, 2008) and are receiving increased attention in many application areas (Candès et al., 2006; Figueiredo et al., 2007; Beck and Teboulle, 2009; Wu et al., 2009). Sparse learning has also been applied in neuroimaging to study genetic influences on the brain (Vounou et al., 2010; Hibar et al., 2011; Kohannim et al., 2011; Le Floch et al., 2011; Vounou et al., 2012; Wang et al., 2012a), functional connectivity (Huang et al., 2010; Ryali et al., 2012), and for outcome predictions (Sun et al., 2009a; Shen et al., 2010; Stonnington et al., 2010; Wang et al., 2010a; Wang et al., 2010b; Wang et al., 2011a). In many computer vision, medical imaging and bioinformatics applications, using sparsity as a prior leads to state-of-the-art results (Sun et al., 2009a; Wright et al., 2009; Liu and Ye, 2010a; Liu et al., 2010b).
Here we developed a new approach, based on conformal slit mapping (Wang et al., 2009a), multivariate tensor-based morphometry (mTBM), and sparse learning, to identify cortical biomarkers for classification problems. We hypothesized that mTBM might improve the accuracy for analyzing group differences in neuroimaging data, and for helping individual classification, when used with a sparse learning classifier. We tested our hypothesis on a dataset used in a prior work (Thompson et al., 2005): it consists of 42 subjects with genetically confirmed William syndrome and 40 age-matched controls. The point of using Williams syndrome data as a test is that the diagnosis can be verified using a genetic test. Despite many years of research on brain differences in Williams syndrome - finding differences widely distributed in the brain - no one known trait offers powerful group classification on its own. As such, we chose this dataset as an interesting test case, as it may also identify distinctive cortical features for further study.
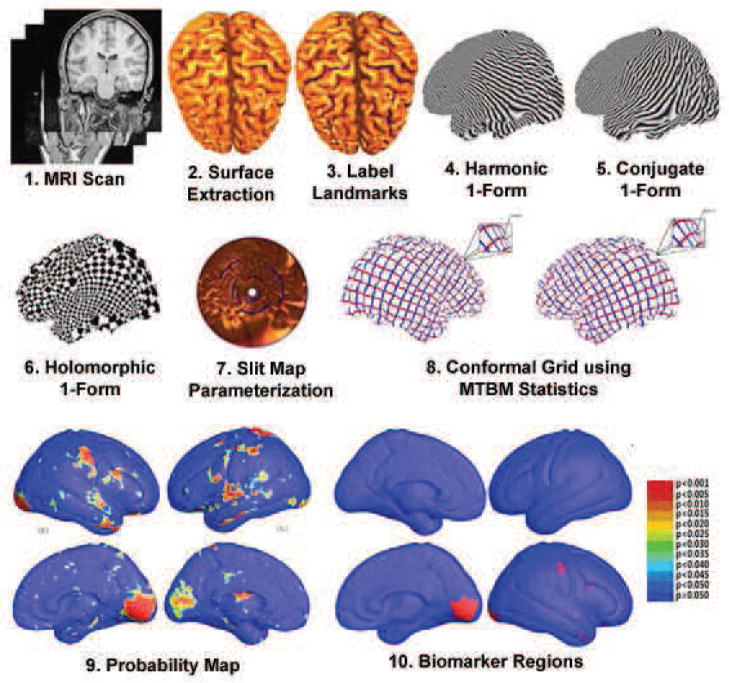
Fig. 1 summarizes the steps we used to analyze cortical surface morphometry. The cortical surface data was from our prior study (Thompson et al., 2005). With 10 selected landmarks on each cortical hemispheric surface, we computed a conformal mapping from a multiply connected mesh to the so-called slit domain, which consists of a canonical rectangle or disk in which 3D curved landmarks on the original surfaces are mapped to parallel lines or concentric slits in the slit domain (Wang et al., 2008a). In this canonical parametric domain, cortical surfaces were matched by a constrained harmonic map (Wang et al., 2007). Multivariate surface statistics were computed from the registered surfaces (Wang et al., 2010c). In one experiment, they were applied to identify regions with significant differences between the two groups. In another experiment, cortical features were fed to a sparse learning method to classify each subject into one of two groups by a leave-one-out test. We also tested other possible surface morphometry statistics to compare them with our multivariate surface statistics. Although the method is illustrated on Williams syndrome data, it is intended to be useful for other disorders as well. Tests on more diverse datasets are reserved for further work.
Figure 1.
A flow chart shows how circular slit map conformal parameterization is used to model cortical surface shapes. The resulting surfaces are analyzed using multivariate tensor-based morphometry and sparse learning methods. After cortical surfaces are extracted from MRI images and landmark curves are labeled either manually or automatically (Thompson et al., 2005), we compute circular slit map conformal parameterizations for each cortical surface, and register surfaces with a constrained harmonic map. The statistics of multivariate TBM are computed at each point on the resulting matching surfaces, revealing regions with systematic anatomical differences between groups. We also apply a sparse learning algorithm to detect some structural features suitable for classification experiments.
2. SUBJECTS AND METHODS
2.1. Subjects
We tested our algorithm on data from a prior study by Thompson et al. (2005). Subjects and brain-scanning protocols were used exactly as in the study by (Reiss et al., 2004; Thompson et al., 2005). Exclusion criteria included a history of medical conditions not typically associated with WS, such as epilepsy or other neurological conditions. All WS participants were evaluated at the Salk Institute (La Jolla, CA) as part of a program project on genetics, neuroanatomy, neurophysiology, and cognition. WS diagnosis was genetically confirmed in all cases by fluorescent in situ hybridization, which tested for deletion of one copy of the elastin gene on chromosome 7. A total of 42 subjects with genetically confirmed Williams syndrome and 40 age-matched healthy controls were included in the study. The studying subject age and sex information is listed in Table 1. Wechsler Full-Scale intelligence quotient (IQ) scores were available for 41 of the 42 WS subjects (mean, 68 ± 9; range, 46–83); the untested subject exhibited similar levels of cognitive function on other measures. As in the earlier studies (Reiss et al., 2004; Thompson et al., 2005), healthy control subjects (with no history of major psychiatric, neurological, or cognitive impairment) were recruited at both the Salk Institute and Stanford University. Control subjects were further screened to rule out any history of learning, language, or behavioral disorder. The majority of controls in the study did not have IQ testing performed. Those that did (n = 16) had a mean full-scale IQ of 104 with an SD of 12 (range, 86 – 126). All procedures were approved by the Institutional Review Boards of both institutions, and all participants provided informed consent (and parents or guardians provided written consent where appropriate). 3D MRI brain images were collected using a GE-Signa 1.5T scanner (General Electric, Milwaukee, MI). The same 3D spoiled gradient echo pulse sequence was used for all participants, with the following parameters: echo time, 5ms; repetition time, 24 ms; flip angle, 45° ; number of excitations, 2; matrix size, 256 × 192; field of view, 24cm; slice thickness, 1.2 mm; 124 contiguous. Scans were analyzed at the University of California Los Angeles Laboratory of Neuroimaging by image analysts blinded to all subject information, including age, gender, and diagnosis (Thompson et al., 2005). All MR images were processed with a series of manual and automated procedures to build brain cortical surfaces and trace landmark curves. The procedure has been described in detail in earlier studies (Thompson et al., 2003; Thompson et al., 2004b; Thompson et al., 2005). In order to extract the brain surfaces, first, non-brain tissue (i.e., scalp, orbits) was removed from the images, and each image volume was re-sliced into a standard orientation by a trained operator who “tagged” 20 standardized anatomical landmarks in each subject’s image dataset that corresponded to the same 20 anatomical landmarks defined on the International Consortium for Brain Mapping-53 average brain (Mazziotta et al., 2001; Thompson et al., 2003; Thompson et al., 2005). Next, a least-squares, rigid-body transformation spatially matched each individual to the average of the healthy control group. In this way, every individual’s brain was matched in space, but global differences in brain size and shape remained intact. Automated tissue segmentation was conducted for each volume dataset to classify voxels as most representative of gray matter, white matter, cerebrospinal fluid (CSF), or a background class (representing extra-cerebral voxels in the image) on the basis of signal intensity. The procedure fits a mixture of Gaussian distributions to the intensities in each image before assigning each voxel to the class with the highest probability (Shattuck et al., 2001). Then each individual’s cortical surface was extracted and three-dimensionally rendered using automated software (MacDonald, 1998). Each resulting cortical surface was represented as a high-resolution mesh of 131,072 surface triangles spanning 65,536 surface points (Thompson et al., 2005). An image analyst, blind to subject diagnosis, gender, and age, traced each of 10 sulci in each hemisphere on each hemisphere surface (Sowell et al., 2003) rendering of each subject’s brain*.
Table 1.
Subject age, sex information within groups.
| Patients of WS (n=42) | Control (n=40) | |
|---|---|---|
| Age mean ± SD | 29.2 ± 9.0 | 27.5 ± 7.4 |
| Age range | 12–50 | 18–49 |
| Male/female | 19/23 | 16/24 |
2.2. Overview of Surface Conformal Slit Mapping
To compute the multivariate statistics, the images need to be registered by an accurate and efficient method, as is also true in the case of more standard morphometric mapping methods, such as voxel based morphometry (VBM) (Ashburner and Friston, 2000) and tensor based morphometry (TBM) (Hua et al., 2012). Thus the first step in our system is to register the brain surfaces from different subjects onto a common template to establish diffeomorphic (one-to-one, smooth) cross-subject correspondences. The surface parameterization process allows us to compare and analyze surface data effectively on a simpler parameter domain, instead of considering the complicated surfaces. A key direction in brain surface registration is to compute a canonical parameter space that can maintain certain properties of the 3D surface, such as geodesics and angles. The conformal slit map (Wang et al., 2008a) method is a numerically stable and efficient method that can induce parametric grids onto surfaces with complicated topologies (multiple boundaries). In this work, we chose slit mapping to construct the canonical parameter space for surface registration. In the Appendix, we briefly explain the mathematical background and implementation details of surface conformal slit mapping.
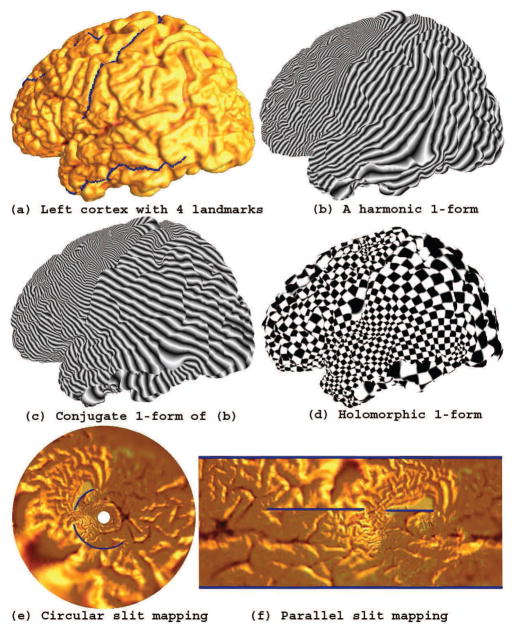
Fig. 2 illustrates the procedure to compute conformal slit mapping on a left hemisphere cortical surface. First, we converted the cortical surface to an open boundary surface by cutting along 4 specific landmark curves. Fig. 2 (a) shows the landmark curves labeled by blue color (the last landmark curve is on the back so it is invisible here). (b) shows a computed exact harmonic 1-form. (c) shows the conjugate 1-form of the exact harmonic 1-form in (b). The final conformal parameterization is illustrated by the texture mapping of the check board image (d). Its circular conformal slit mapping result is shown in (e), where two landmarks were mapped to inner and outer circles while the other two were mapped to two concentric slits. The parallel conformal slit mapping result is shown in (f), where the four landmark curves were mapped to the blue straight lines.
Figure 2. Conformal slit mapping.
After cutting along several landmark curves, we turn a cortical surface to a genus zero open boundary surface (a). By computing the holomorphic 1-form (d), this may then be conformally mapped onto either a circle (e) or a rectangle (f) where the landmarks are mapped to concentric or parallel lines in the slit domain. We then perform nonlinear surface registration in these parameter domains.
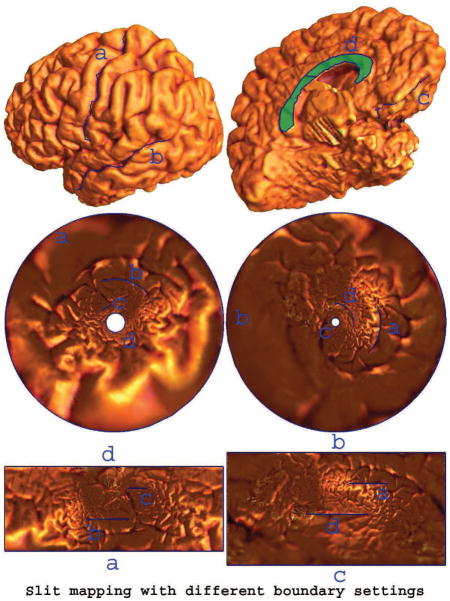
The slit mapping method conformally maps a multiply connected surface to an annulus or a strip with two parallel boundaries. The choice of which boundary is outer is flexible. This flexibility may be useful for selection of the most meaningful conformal parameterization for certain applications. Fig. 3 illustrates a conformal slit mapping result of a left hemisphere cortical surface with 4 boundaries. We selected 5 landmark curves with linking Callosal Sulcus and Inferior Callosal Outline Segment. We cut along the selected landmarks to obtain a genus zero surface with 4 boundaries. In Fig. 3, we show both circular and parallel slit mapping parameterization results while selecting two different pairs of landmark curves as the boundaries in the parameter domains.
Figure 3. Slit mapping with different boundary settings.
For a given cortical surface, we cut it open along 5 selected landmark curves and thus the surface becomes a genus zero surface with 4 open boundaries (first row). The second row shows two circular slit map conformal parameterization results with different landmark curves as outer and inner circular boundaries. The third row shows their parallel slit map conformal parameterization results with different landmark curves as the outer boundaries.
2.3. Surface Registration by Constrained Harmonic Map
Brain surface deformation studies typically require the computation of dense correspondence vector fields to match one brain surface with another. Many brain surface registration methods have been proposed (Fischl et al., 1999; Thompson et al., 2000b; Van Essen et al., 2001; Memoli et al., 2004; Vaillant and Glaunes, 2005; Wang et al., 2005; Qiu and Miller, 2007; Durrleman et al., 2008; Tosun and Prince, 2008; Auzias et al., 2009; Pantazis et al., 2010; Yeo et al., 2010; Zhong and Qiu, 2010). Here we propose to use the slit mapping method to generate a canonical surface to match brain surfaces. Conformal structure is intrinsic to surfaces. As landmark curves have similar geometric positions on different cortical surfaces, the conformal structures of the converted multiply connected components are relatively similar. As a result, their conformal parameterizations to the slit domain are similar to each other.
We propose to compute a direct correspondence between any two surfaces by solving a constrained harmonic map problem (Wang et al., 2007). A harmonic map is one of the most broadly applied methods for registration of computational models of surfaces (Gu et al., 2004; Joshi et al., 2007; Wang et al., 2007). Among its many advantages, the harmonic map (1) measures the elastic energy of the deformation, so it has a clear physical interpretation; (2) can be computed by solving an elliptic partial differential equation, so its computation is numerically stable; and (3) it continuously depends on the boundary condition, so it can be controlled by adjusting the boundary condition. Given two surfaces S1 and S2, their conformal slit mappings are τ1 : S1 → ℝ2 and τ2 : S2 → ℝ2. We want to compute a map φ : S1 → S2. Instead of a direct computation of φ, we can easily find a harmonic map between their parameter domains, i.e. we look for a harmonic map τ: ℝ2 → ℝ2, such that
| (1) |
Then the map φ can be obtained by . Since τ is a harmonic map while τ1 and τ2 are conformal maps, the resulting φ is a harmonic map. With the constrained harmonic map in the slit parameter domain, we are able to obtain exact landmark matching across subjects, which may boost the power of the multivariate statistics.
2.4. Multivariate Tensor-Based Morphometry (mTBM)
To study structural features of the brain, such as cortical gray matter thickness, complexity, and deformation over time, there are two basic approaches: deformation-based morphometry (DBM) (Ashburner et al., 1998; Chung et al., 2001; Chung et al., 2003; Wang et al., 2003) and tensor-based morphometry (TBM) (Davatzikos et al., 1996; Thompson et al., 2000a; Chung et al., 2008). DBM tends to analyze 3D displacement vector fields encoding relative positional differences across subjects, while TBM tends to examine spatial derivatives of the deformation maps registering brains to a common template, constructing morphological tensor maps such as the Jacobian determinant, strain, torsion, or even vorticity.
We used multivariate statistics based on the surface deformation tensors to study cortical surface morphometry (Wang et al., 2010c; Wang et al., 2011b). Suppose φ : S1 → S2 is a map from surface S1 to surface S2. The derivative map of φ is the linear map between the tangent spaces dφ : TM(p) → TM(φ(p)), induced by the map φ, which also defines the Jacobian matrix of φ. In the triangle mesh surface, the derivative map dφ is approximated by the linear map from one face [v1, v2, v3] to another [w1, w2, w3]. First, we isometrically embed the triangles [v1, v2, v3] and [w1, w2, w3] onto the plane ℝ2; the planar coordinates of the vertices of vi, wj are denoted using the same symbols vi, wj. Then we explicitly compute the Jacobian matrix for the derivative map dφ (Wang et al., 2008b).
| (2) |
In our work, we use multivariate statistics on deformation tensors (Brun et al., 2008; Leporé et al., 2008) and adapt the concept to surface tensors. Let J be the Jacobian matrix and define the deformation tensors as . Instead of analyzing shape change based on the eigenvalues of the deformation tensor, we consider a new family of metrics, the “Log-Euclidean metrics” (Arsigny et al., 2006). These metrics make computations on tensors easier to perform, as the transformed values form a vector space, and statistical parameters can then be computed easily using standard formulae for Euclidean spaces (Wang et al., 2008b).
To compute group differences with mTBM, we propose to apply Hotelling’s T2 test (Hotelling, 1931; Cao and Worsley, 1999; Thirion et al., 2000; Kim et al., 2012) on sets of values in the log-Euclidean space of the deformation tensors. Given two groups of n × 1-dimensional vectors, Si, i = 1,2, …, p, Tj, j = 1,2, …, q, we use the Mahalanobis distance M to measure the group mean difference,
| (3) |
where S̄ and T̄ are the means of the two groups and Σ is the combined covariance matrix of the two groups (Leporé et al., 2008; Wang et al., 2010c; Wang et al., 2011b). In our study, S and T are the log-Euclidean metrics, e.g. , i = 1,2, …, p and , j = 1,2, …, q.
2.5. Structural Classification Feature Detection with Stability Selection
We used mTBM to detect structural features for group classification (here, i.e. WS vs. controls). For a classification algorithm based on 3D images or surface-based features, the feature dimension is usually much larger than the number of subjects in the sample, so some feature reduction is necessary. There are many feature selection approaches in the data mining literature (Kuncheva and Rodríguez, 2010). One unique challenge for mTBM based feature detection on brain surfaces is the huge dimensionality of the dataset. Therefore, feature selection is performed before the classification process.
Traditionally, feature selection methods, e.g., the recursive feature evaluation method (Guyon et al., 2002; Fan et al., 2005), Sequential Forward Selection (SFS) (Stearns, 1976), are largely based on the discriminative power of each individual variable. However, with huge dimensionality, a feature can gain statistical significance just by pure chance, without having any generalizability to new unseen datasets. In addition, many feature selection algorithms tend to select a vast number of features, which may have no biologically plausible interpretation. To address this problem, we used sparse learning to identify useful features. Sparse signal representation has proven to be an extremely powerful tool for acquiring, representing, and compressing high-dimensional signals (Candès and Tao, 2005; Bruckstein et al., 2009).
For our problem, we consider the dataset d = {(ai, yi)|i = 1,2, …, n}, in which ai is the feature vector, y = ±1 is the class label, and n is the number of samples. For a linear classifier h = aw, we can learn it by solving the following optimization problem:
| (4) |
This procedure is also called the “Lasso” (Tibshirani, 1996), in which λ is the regularization term that balances between sparsity and training error. The only difference between the lasso and the well-known ridge regression (Tikhonov and Arsenin, 1977; Kohannim et al., 2011) is the second term. The ℓ1-norm regularization will induce an interesting property, i.e., in the solution of the lasso problem, a portion of w will be exactly zero. This provides us a method that will jointly select useful features, and this can be adapted to our high-dimensional problem. After feature selection, the dataset is reduced to a reasonable size and classification is performed. In this stage, we apply ridge regression (Tikhonov and Arsenin, 1977; Kohannim et al., 2011) to discriminate between individuals with WS and healthy controls.
We employed Nesterov’s Accelerated Gradient Method (AGM) to solve the proposed formulation (Eq. 4) (Nesterov, 2007). AGM achieves the optimal convergence rate among all first-order methods (Nesterov, 2007). The core computation for AGM is the proximal operator associated with different regularization Ω(·) (Combettes and Pesquet, 2009). We have developed efficient algorithms to compute the proximal operators in our software package, SLEP (Sparse Learning with Efficient Projections) (Liu et al., 2009). SLEP achieves state-of-the-art performance for many sparse learning models (currently, the package has around 4,000 active users from 17 different countries). The SLEP tools have been applied successfully for stroke prediction (Khosla et al., 2010), craniosynostosis classification (Yang et al., 2011), joint gene expression and network data analysis (Ji et al., 2009; Liu and Ye, 2010b), to predict conversion from mild cognitive impairment (MCI) to AD (Ye et al., 2012), and for brain network analysis in Alzheimer’s disease (Sun et al., 2009b).
To validate the generalization of our model and selected biomarker sets, we changed registering template surfaces and chose stability selection (Meinshausen and Bühlmann, 2010) to select the highest ranking features. Stability selection is a bootstrapping approach to control for false discoveries in the case of finite samples and has been specifically proposed for sparse predictive modeling (Meinshausen and Bühlmann, 2010). The key to stability selection is to perturb the data many times and choose features that occur in a large fraction of the resulting selection sets. Thus, choosing the right value of the regularization parameter λ becomes much less critical and we have a better chance of selecting truly relevant features.
In our experiments, for each feature, we maintain a counting number and put its initial value as 0. The bootstrapping is done on random subsamples of {1, …, n} of size n/2, drawn without replacement. In each step, we solve Eq. 4 and obtain a sparse solution. If a feature is selected, we increase its counting number by 1. After we repeat this procedure s times (e.g. 10,000 times), we obtain a counting number, cxgi for each feature. We define its selection probability as . For a cut-off πthr(0 < πthr < 1) and a set of regularization parameters, Λ, the set Ŝstable of stable features k is defined by:
| (5) |
Using this method, we can determine a set of statistical significant features that may be either used for classification or for visualization of some stable feature sets to explain anatomical differences. The stability selection (Meinshausen and Bühlmann, 2010) is appealing in that it has strong theoretical guarantees. Specifically, it has been shown that subsampling/bootstrapping in conjunction with ℓ1-regularized estimation requires much weaker assumptions on the data for asymptotically consistent feature selection than what is needed for the traditional ℓ1-regularized scheme. It has been proven to improve estimation of discriminative features significantly, even in cases when the necessary conditions for consistency of the original Lasso method are violated. It has been adopted by some recent brain imaging research work (Ryali et al., 2012; Vounou et al., 2012; Ye et al., 2012).
3. RESULTS
We tested our method on cortical surface data from 42 subjects with genetically confirmed WS and 40 age-matched healthy controls (Thompson et al., 2005). With the slit map method, each cortical surface was conformally mapped to an annulus with concentric arcs. We then computed a constrained harmonic map to register the surfaces to a template surface, using Eq. 1. The template surface was chosen randomly from the control set. The constrained harmonic map helped us build a direct correspondence between the cortical surfaces.
Cortical surface models were cut along 10 landmark curves per hemisphere: the Central Sulcus, Superior Temporal Sulcus (Main Body), Inferior Frontal Sulcus, Middle Frontal Sulcus, Inferior Temporal Sulcus, Secondary Intermediate Sulcus, Transverse Occipital Sulcus, Inferior Callosal Outline Segment, Superior Rostral Sulcus, and Subparietal Sulcus. Landmarks were defined according to a detailed anatomical protocol (Sowell et al., 2002) based on the Ono sulcal atlas (Ono et al., 1990). The written anatomical protocol is available on the internet (Hayashi et al., 2002) and has been used in many published studies. After we cut each cortical surface open along the selected landmark curves, the cortical surface became topologically equivalent to a genus zero surface with 10 boundaries (10 landmark curves). By computing a sequence of differential geometric features on these surfaces - exact harmonic 1-forms, closed harmonic 1-forms, and holomorphic 1-forms - we built a circular slit map, which conformally mapped each surface onto an annulus with 10 boundaries (similar to Fig. 2 (e)). We then matched all surfaces by the constrained harmonic map (Wang et al., 2007). To study surface morphometry, the Jacobian matrices were computed as Eq. 2.
Group Difference Results
We used the Hotelling’s T2 test (Hotelling, 1931) to identify between-group differences. Specifically, for each point on the cortical surface, given p=0.05 as the significance level, we ran a permutation test with 10,000 random assignments of subjects to groups to estimate the statistical significance of the areas with group differences in surface morphometry. Fig. 4 shows the significance map of group differences detected between WS and matched control groups, using mTBM as a measure of local surface area and the significance at each surface point to be p=0.05. In Fig. 4, the non-blue colored areas show the areas with (uncorrected) statistically significant differences between the two groups.
Figure 4.
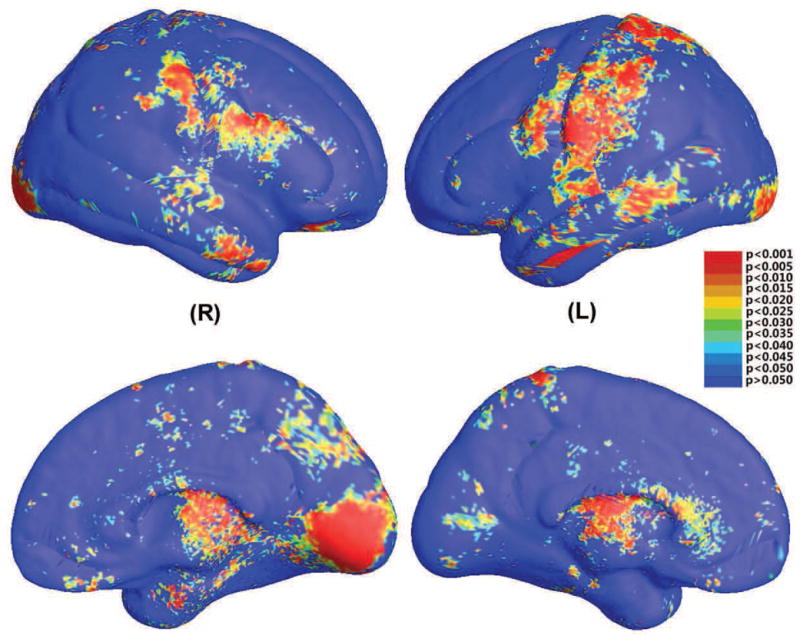
Statistical significance map (uncorrected p-map) shows group differences in regional cortical surface area between 42 WS patients and 40 healthy controls (Thompson et al., 2005). The local statistic analyzed is the multivariate TBM of the cortical parameterization. On the color-coded scale, non-blue colors denote the vertices where there are significant group differences, at the uncorrected p=0.05 level.
The overall significance of the map can be defined as the probability of finding, by chance alone, a statistical map with at least as large a surface area and a statistical threshold more stringent than the predefined level of p=0.05 (note that other methods are also possible, such as those that control the false discovery rate). This omnibus p-value is commonly referred to as the overall significance of the map (or of the features in the map), corrected for multiple comparisons. It basically quantifies the level of surprise in seeing a map with this amount of the surface exceeding a predefined threshold, under the null hypothesis of no systematic group differences. We also computed the overall significance p-values, which were p=0.0004 for the left and p=0.001 for the right hemisphere, respectively.
Classification Results
After we computed the Log-Euclidean metrics to establish a metric on the surface deformation tensors at each point, we applied the ℓ1-norm penalty (Eq. 4) as the loss function to select sparse features for classification. We used the SLEP package (Liu et al., 2009) for the computation.
In our experiments, we took a cross validation approach to set up free parameters in the Lasso (λ in Eq. 4) and ridge regression method (λ). In our new experiments, in each round of experiments, we divided the training data into four equally sized subsets. Among them, we did cross validation to select the free parameters. Specifically, with a set of selected free parameter values with Lasso and rigid regression classifier, we used three subsets for training and tested the trained classifiers on the last subset. Continue this procedure, each time omitting one subset from the training data and then testing on the omitted one part from the training data and testing on the omitted part. Then the combined 4 part results are put together to estimate the total error rates for the performance comparisons of the free parameter values. We continuously tried all combinations of free parameters and selected the free parameter values by choosing the ones which achieved the best total error rate on the training dataset. Then we applied the chose regularization parameter set to test on the testing dataset.
A leave-one-out method was used to evaluate our classification. First, we randomly removed a sample from the dataset, and set up the free parameter values with a cross validation on the remaining set. From the solution of the Lasso problem, we selected non-zero entries as the useful features. Using only these features, we applied ridge regression to obtain the classifier. Next, the previously removed sample was examined by the classifier. The experiment continued in this leave-one-out fashion, and overall annotation accuracy was calculated at the end. This procedure was repeated 82 times, each time with a different choice for the test data. During the prediction of the ridge regression classifier, instead of the immediate label, a test sample was assigned a score, s, where a positive s value was assigned as WS class or to control (CTL, or typically developing) class. By observing the distribution of the scores for both classes, we can obtain an intuitive impression of the classification power.
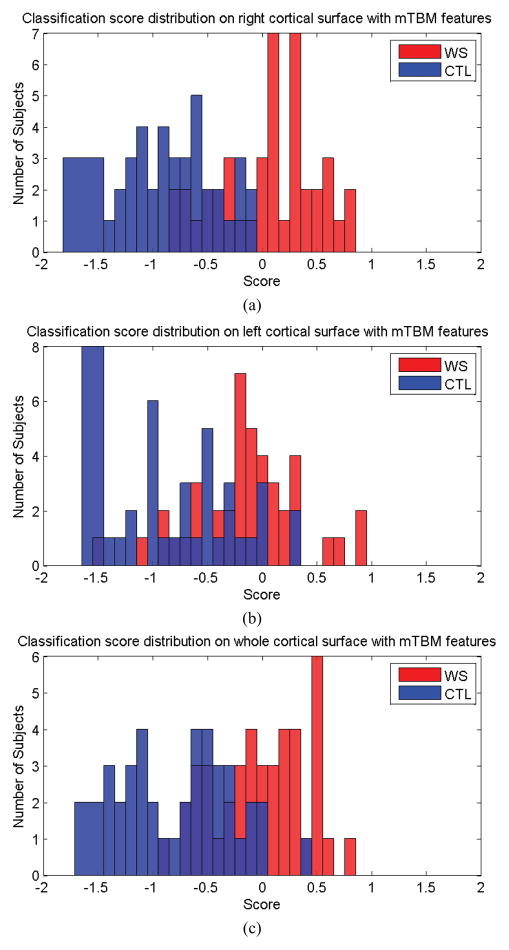
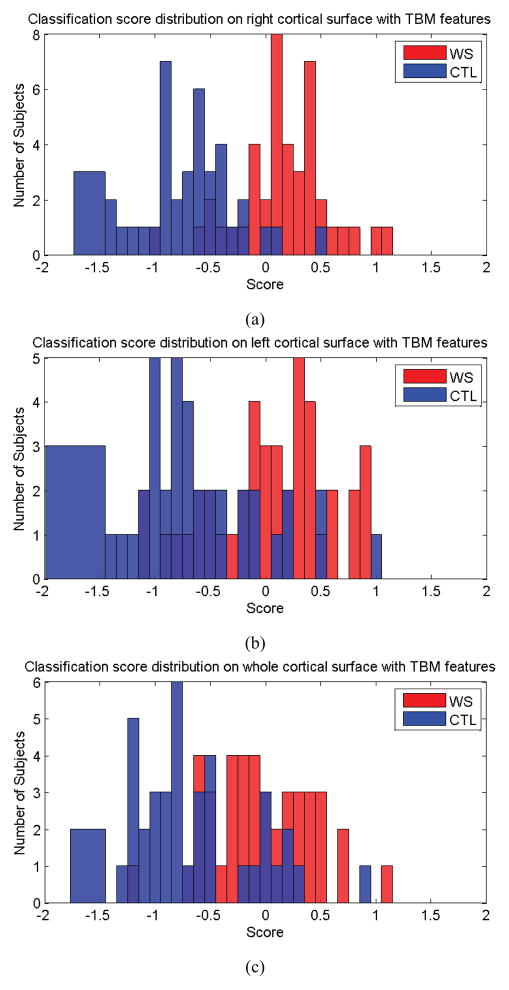
In a series of experiments, the 82 results were used to compute overall performance measures. Fig. 5 shows the classification scores for each subject in the prediction test. Fig. 5 (a) shows the scores for each subject with mTBM features only, on the right cortical hemisphere; in this case, all control subjects were correctly classified while 17 WS patients were incorrectly classified as control subjects. Fig. 5 (b) and (c) show the scores on each subject, with mTBM on the left cortical hemisphere, and using both the left and right cortical hemispheres, respectively.
Figure 5.
Classification scores obtained for both classes, using mTBM on the right hemisphere (a), left hemisphere (b), both left and right hemispheres (c).
The output of each classifier was compared to the ground truth, and a contingency table was computed to indicate the number of particular class labels that were correctly identified as members of one of two classes. The rows of the contingency table represent the true classes and the columns represent the assigned classes. The cell at row r and column c is the number of subjects whose true class is r while their assigned class is c. The possible true- and detected-state combination is shown in Table 2. Four performance measures Sensitivity, Specificity, Positive predictive value, and Negative predictive value were computed as follows:
Table 2.
Possible combinations of ground truth and predicted classifications for two classes. This matrix is used to compute the classifier’s accuracy.
| True Class | Assigned Class | |
|---|---|---|
| WS | CTL | |
| WS | Nww | Nwc |
| CTL | Ncw | Ncc |
The contingency table for mTBM features is shown in Table 3. Its performance measures are shown in rows 2–4 of Table 8. We can find that the best specificity (100.00%), sensitivity (59.52%), positive predictive value (100.00%) and negative predictive value (70.18%) were achieved when we used mTBM features from the right cortical hemisphere for the training and testing. Consistent with some data from prior WS studies (Thompson et al., 2005), the right half of the brain may contain more diagnostically useful information relevant to WS classification. This assumption is supported by our classification results.
Table 3.
Classification performance with mTBM features on right cortical hemisphere, left cortical hemisphere and both cortical hemispheres.
| mTBM on right cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 25 | 17 | |
| CTL | 0 | 40 | |
| mTBM on left cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 15 | 27 | |
| CTL | 2 | 38 | |
| mTBM on both cortical hemispheres | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 21 | 21 | |
| CTL | 1 | 39 | |
Table 8.
Classification experimental result comparison with various feature groups.
| Sensitivity(%) | Specificity(%) | Positive Value(%) | Negative Value(%) | |
|---|---|---|---|---|
| mTBM on right cortical hemisphere | 59.52% | 100.00% | 100.00% | 70.18% |
| mTBM on left cortical hemisphere | 35.71% | 95.00% | 88.24% | 58.46% |
| mTBM on both cortical hemispheres | 50.00% | 97.50% | 95.45% | 65.00% |
| TBM on right cortical hemisphere | 71.43% | 92.50% | 90.91% | 75.51% |
| TBM on left cortical hemisphere | 52.38% | 85.00% | 78.57% | 62.96% |
| TBM on both cortical hemispheres | 42.86% | 85.00% | 75.00% | 58.62% |
| Pair of EV of J on right cortical hemisphere | 52.38% | 92.50% | 88.00% | 64.91% |
| Pair of EV of J on left cortical hemisphere | 50.00% | 87.50% | 80.77% | 62.50% |
| Pair of EV of J on both cortical hemispheres | 42.86% | 90.00% | 81.82% | 60.00% |
| Volume of both cortical hemispheres | 61.90% | 52.50% | 57.78% | 56.76% |
Stability Selection for Verification of Feature Sets
In our leave-one-out experiments, we randomly selected a subject as the template and used it to register other surfaces. We used this approach in our prior work (Wang et al., 2009a; Wang et al., 2012b). Our earlier work (Wang et al., 2009b) indicated that it could give rise to different performances to use different surfaces as the registering template. In Wang et al. (2009b), we used surface Ricci flow method to conformally parameterize cortical surfaces. We proposed to use conformal invariants to rank template surfaces and our empirical results showed that it may provide some assistance for template surface selection (Wang et al., 2009b). However, in general it is still challenging to address the optimal registering template selection problem because of the variety of anatomical surfaces. The problem became even more elusive here since conformal invariants proposed in (Wang et al., 2009b) are not computable when we use conformal slit mapping to parameterize cortical surfaces.
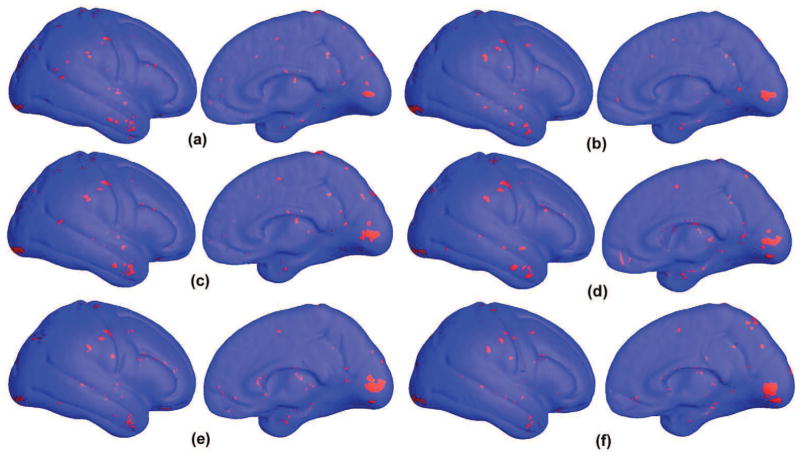
To visualize the selected feature set with different registering template surfaces, here we propose to apply the stability selection to select top features when we use six different cortical surfaces as template for surface registration. Fig. 6 shows the selected top features in these six experiments. The red pixels are selected by the stability selection method as significant features for group discrimination and diagnostic classification at the individual level: a total of 405, 429, 453, 451, 426 and 452 vertices selected in (a), (b), (c), (d), (e) and (f), respectively (πthr=0.25); (a) is the result with the template surface we used in our leave-one-out cross validation experiments. Since all surfaces are used by the stability selection method, they are not the feature set used in each of the 82 leave-one-out experiments. However, they are a set of statistical significant features that may be used for visualization to help explain anatomical differences. As shown in Fig. 6, the selected vertices are consistent among the six feature sets. This suggests that our method is relatively robust and independent of the particular selection of template surface for surface registration. Although how much a template surface in our surface registration framework affects the final classification results requires careful validation for each application, the current results show our method can select stable feature sets for classification.
Figure 6.
Feature vertices selected by the stability selection method (Meinshausen and Bühlmann, 2010) on the average right hemisphere with three different template cortical surfaces used in the registration step. The red pixels are selected by the stability selection method as significant features for group discrimination and diagnostic classification at the individual level: a total of 405, 429, 453, 451, 426 and 452 vertices selected in (a), (b) (c), (d), (e) and (f), respectively (πthr =0.25). The selected vertices are relatively consistent among three feature sets. Although more careful validation is needed to explore how the template surface selection affects the final classification results, the current results show our method can select stable feature sets for classification.
Group Difference Comparison between mTBM and Other Surface Morphometry Statistics
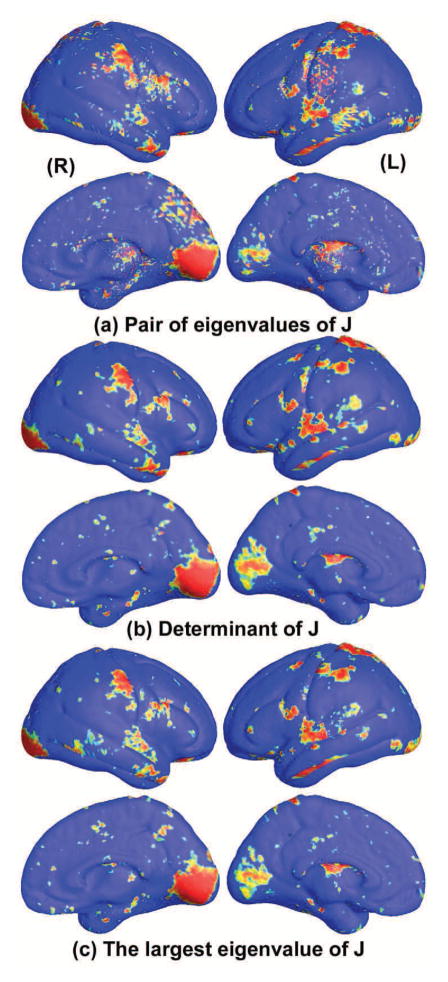
To test the power of multivariate TBM statistics, we also conducted three additional statistical tests based on different tensor-based statistics derived from the Jacobian matrix. The other statistical tests evaluated were: (1) the pair of eigenvalues (EV) of Jacobian (J) matrix, treated as a 2-dimesional vector; (2) the determinant of Jacobian matrix; (3) the largest eigenvalue of Jacobian matrix. For statistics (2) and (3), we applied a Student’s t test to compute the group mean difference at each surface point. In case of statistic (1), we used Hotelling’s T2 test to compute the group mean difference. Fig. 7 shows the significance maps of group differences detected between WS and matched control groups, using surface statistics (1)–(3) as a measure of local surface area and the significance at each surface point to be p=0.05. In Fig. 7, the non-blue color areas denoted the statistically significant areas between two groups. In the experiment, all group difference p-maps were corrected using a permutation test with 10,000 random assignments of subjects to groups. In Figs. 4 and 7, we can see that the detected significant areas are consistent for all four statistics. Table 4 gives the permutation corrected p-values comparison.
Figure 7.
Comparison of p-maps for group differences (between Williams syndrome and typically developing participants) with the other three surface statistics.
Table 4.
Permutation test results with four surface statistics.
| mTBM | Pair EV of J | Determinant of J | Largest EV of J | |
|---|---|---|---|---|
| Left hemisphere | 0.0004 | 0.0001 | 0.0006 | 0.0009 |
| Right hemisphere | 0.001 | 0.0003 | 0.0043 | 0.0033 |
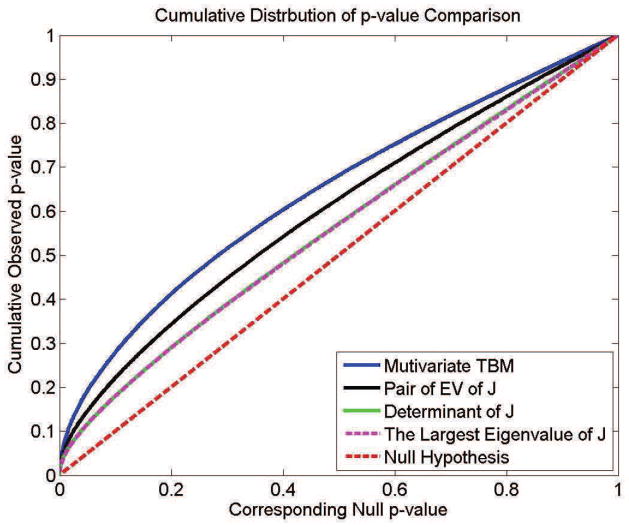
In Fig. 8, the cumulative distribution functions (CDF) of the p-values observed for the contrast of patients versus controls are plotted against the corresponding p-value that would be expected, under the null hypothesis of no group difference, for the different scalar and multivariate statistics. For null distributions, the cumulative distribution of p-values is expected to fall approximately along the line (represented by the dotted line); large deviations from that curve are associated with significant signal, and greater effect sizes represented by larger deviations (the theory of false discovery rates gives formulae for thresholds that control false positives at a known rate). We note that the deviation of the statistics from the null distribution generally increases with the number of parameters included in the multivariate statistics, with statistics on the full tensor typically outperforming scalar summaries of the deformation based on the eigenvalues.
Figure 8.
Comparison of cumulative distribution functions for group differences, using different surface statistics. Based on the FDR method, this type of QQ plot can be used to show if there is aggregate evidence of a signal in a statistical map, across all statistical thresholds.
From Table 4 and Fig. 8, we can see that the mTBM feature (Sec. 2.5) performs better than other two statistics, (2) and (3) - TBM and the largest eigenvalue of Jacobian matrix. Between mTBM and statistic (1), the pair of eigenvalues of Jacobian matrix, we can see that the latter has more significant permutation test results but mTBM has a steeper CDF curve (Fig. 8), i.e. more deviation from the null distribution curve. Considering the pair of eigenvalues is also a kind of multivariate surface statistics, we concluded that generally the multivariate tensor-based morphometry method may be able to detect more surface deformation compared with univariate statistics and produce comparable (or slightly better) results relative to other multivariate statistical approaches.
Classification Performance Comparison with Other Brain Surface features and Volume Features
We also performed additional classification experiments with other features. The features we considered included tensor-based morphometry (TBM) – the determinant of Jacobian matrix (Davatzikos et al., 1996; Thompson et al., 2000a; Woods, 2003; Chung et al., 2008), the pair of eigenvalues of Jacobian matrix and the total volume of both left and right cortical hemispheres. In the first two experiments, we first applied Lasso to select features, and then we used ridge regression to do the classification. To set up the parameters of Lasso and ridge regression, we took the same approach as the one used in mTBM experiments. In each round, we divided the training dataset into four subsets. We performed a cross validation on these four subsets with all possible combinations of the parameter values. The parameter set which achieved the best classification accuracy rates on the training dataset was used as the classifier parameters to classify the last testing dataset. We repeat all the procedure on all data, each time omitting one subset from the training data and then testing on the omitted one part from the training data and testing on the omitted part. Then the final classification results were estimated by combining all leave-one-out experiments. Fig. 9 shows the classification scores for TBM features on right hemisphere (a), left hemisphere (b) and both hemispheres (c). Their contingency tables are shown in Table 5 and their performance metrics are shown in rows 5–7 of Table 8. Fig. 10 shows the classification scores for pair EV of J features on right hemisphere (a), left hemisphere (b) and both hemispheres (c). Its contingency table is shown in Table 6 and its performance metrics are shown in rows 8–10 of Table 8.
Figure 9.
Classification scores obtained for both classes, using TBM features – the determinant of Jacobian matrix (Davatzikos et al., 1996; Thompson et al., 2000a; Woods, 2003; Chung et al., 2008) on the right hemisphere (a), left hemisphere (b), both left and right hemispheres (c).
Table 5.
Classification performance with TBM features (the determinant of Jacobian matrix) on right cortical hemisphere, left cortical hemisphere and both cortical hemispheres.
| TBM on right cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 30 | 12 | |
| CTL | 3 | 37 | |
| TBM on left cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 22 | 20 | |
| CTL | 6 | 34 | |
| TBM on both cortical hemispheres | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 18 | 24 | |
| CTL | 6 | 34 | |
Figure 10.
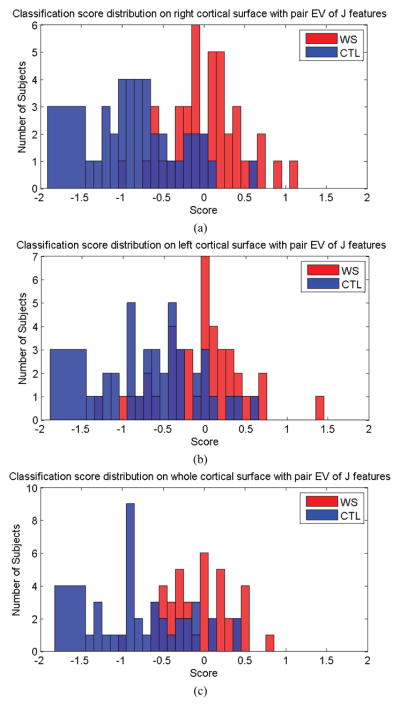
Classification scores obtained for both classes, using the pair of eigenvalues of Jacobian matrix as features for the right hemisphere (a), left hemisphere (b), and for both left and right hemispheres (c).
Fig. 11 shows the classification scores for the cortical volume features. For subject i, we represented its volume as a 2×1 vector, ( ), where and are the volumes of its left and right hemisphere, respectively. Then we directly applied ordinary least squares (Hastie et al., 2001) for the training and classification. Its contingency table is shown in Table 7 and its performance metrics are shown in row 11 of Table 8.
Figure 11.
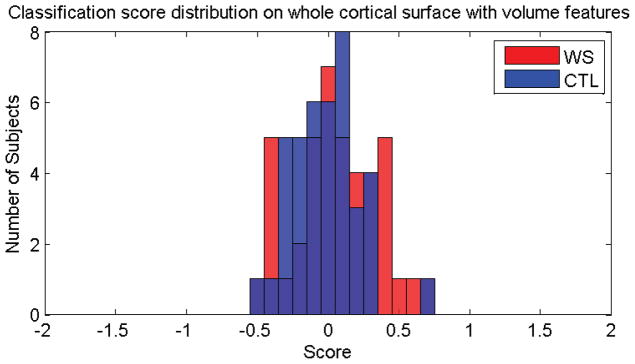
Classification scores obtained for both classes, using the cortical volumes as the features.
Table 7.
Classification performance with total cortical volume features.
| TrueClass | Assigned Class | |
|---|---|---|
| WS | CTL | |
| WS | 26 | 16 |
| CTL | 19 | 21 |
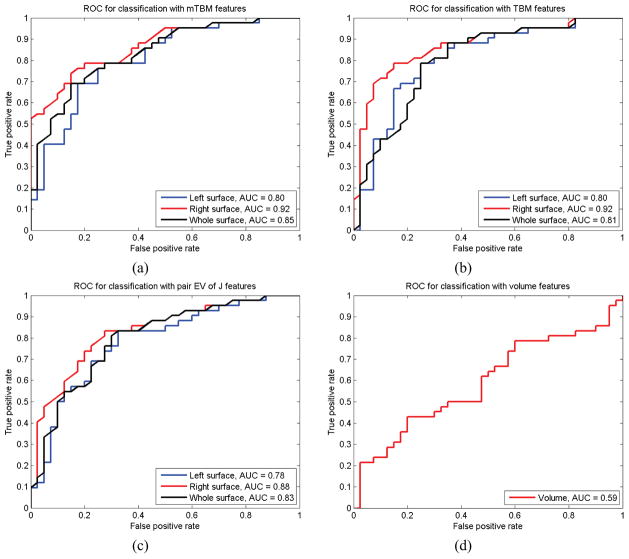
In our experimental results (Table 8), we can find that the best specificity (100%) and positive predictive value (100%) were achieved when we used mTBM features from the right cortical hemisphere for training and testing. The best sensitivity (71.43%) and negative predictive value (75.51%) were achieved when we used TBM features from the right cortical hemisphere for training and testing. To compare performance, we also generated receiver operating characteristic (ROC) curves and computed area-under-the-curve (AUC) measures. In Fig. 12, for the right cortical hemisphere, both mTBM and TBM achieved the best AUC measures (92%), which are better than the results from two other statistical features, i.e., the pair of eigen-values of the Jacobian matrix on the right cortical hemisphere (88%) and cortical volume statistics (59%).
Figure 12.
Classification performance comparison with receiver operating characteristic (ROC) curves and area under curve (AUC) measures. (a)–(d) show results for mTBM, TBM, pair eigen values (EV) of Jacobian matrix and cortical volume statistics, respecitively. With the first three statistics, the results using the right cortical hemisphere only are better than those with left cortical hemisphere and both cortical hemispheres. Among all AUC measures, mTBM on right hemisphere achieved the best performance (AUC=0.94).
4. DISCUSSION
In this paper, our overarching goal was to detect differences in brain surface morphometry. One traditional way to do this is to set up parametric grids on surfaces, and then use differential geometry to come up with useful descriptors of surface features of interest, or to summarize the geometry as a whole. Conformal maps help to induce particularly well-organized grids on surfaces. This simplifies a number of downstream computations of derivatives and metrics. In addition, the surface metric tensor, which is computable from the conformal grid, has a multivariate structure that contains a great deal of information on local surface geometry. Its components cannot simply be inserted into a standard Euclidean multivariate statistical test, as the dependencies among the tensor parameters follow a log-Euclidean law that affects their possible range of values and their statistical distributions. As a result, we end up having to employ the log transform on a matrix, or - more precisely - on a field of matrices or tensors that are defined at the coordinates of the surface grid. The resulting set of surface tensor methods practically encodes a great deal of information that would otherwise be inaccessible, or overlooked. The empirical question is then to see if this extra information is helpful for classification of disease, and if so, under what circumstances.
Our study has two main findings. First, it is possible to detect the subtle difference between or classify people with high accuracy into diseased versus healthy control groups by analyzing surface deformation tensors computed from a set of parametric surfaces using concepts from a limited number of conformal parameterizations. Our current method enforces an exact landmark curve matching on the cortex, using topology optimization. Compared to other cortical surface registration methods based on conformal parameterization, our method eliminates all singular points in the parameterization domains. The analysis of parametric meshes for computational studies of cortical structures can be made more powerful by analyzing the multivariate information inherent in the surface. This type of improvement was shown in group difference analyses. Second, our current experimental results show ℓ1-norm penalty (sparse learning) methods - that use sparsity as a prior - lead to reasonable classification results. Stability selection method is used to visualize the consistency of selected feature points. The sparsity induced by sparse learning methods may help us to account for biological differences using efficient sets of predictors.
We validated our proposed work in a dataset used in our prior work (Thompson et al., 2005). Our earlier work (Thompson et al., 2005) focused on local surface-based cortical thickness differences, while here we explored more complex surface descriptors related to local surface area and the metric tensor. Although these two approaches might be expected to be highly related, in fact – and perhaps surprisingly – recent work by Winkler and colleagues (2010) shows that the genetic influences on cortical surface thickness and regional cortical volume (which depends on surface area as well as thickness) are almost completely non-overlapping. Although this is somewhat non-intuitive, the genetic factors that drive differences in relative area and cortical thickness seem to be very different. This was established using a quantitative genetic model that separates common and distinct sources of genetic variance. As a result, some biological processes may preferentially affect cortical thickness without greatly affecting the relative areas of parts of the cortex, and vice versa. Our current study demonstrated that it is possible to use only surface morphometry information to achieve group classification, offering a new method to determine when human diseases are associated with characteristic neuroanatomical variation.
Comparison with Two Other Conformal Parameterization Methods
The work reported here used parametric surface approaches to register brain anatomical surfaces (Davatzikos, 1996; Bakircioglu et al., 1999; Thompson et al., 2000b) and, as such, is related to ongoing research by ourselves and others on conformal mapping of brain surfaces. Brain surface parameterization has been studied intensively (Schwartz et al., 1989; Angenent et al., 1999; Timsari and Leahy, 2000; Gu et al., 2004; Hurdal and Stephenson, 2004; Wang et al., 2006; Wang et al., 2007; Wang et al., 2008b; Wang et al., 2008a; Hurdal and Stephenson, 2009; Wang et al., 2012b). Most brain conformal parameterization methods (Angenent et al., 1999; Gu et al., 2004; Hurdal and Stephenson, 2004; Joshi et al., 2004; Ju et al., 2004; Ju et al., 2005; Hurdal and Stephenson, 2009) can handle the complete brain cortex surface, but cannot deal with cortical surfaces that have boundaries. The holomorphic flow segmentation method (Wang et al., 2007) can match cortical surfaces with boundaries or landmarks, but the resulting maps have singularities, which lead to errors in practical applications. Only the Ricci flow (Wang et al., 2006; Wang et al., 2008b; Wang et al., 2012b) and slit map methods (Wang et al., 2008a) can handle surfaces with complicated topologies (boundaries and landmarks) without singularities. The Ricci flow method is a nonlinear optimization process, which is more time-consuming than the slit map method. The Ricci flow method also has higher requirements for the quality of surface tessellations. The slit map method uses a linear approach, which tends to be more efficient and robust. In this paper, the slit map method (Wang et al., 2008a) is used to conformally map a multiply connected domain to an annulus with multiple concentric arcs (called the circular slit map) or to a rectangle with multiple straight lines (called the parallel slit map). It is a global conformal parameterization method without needing the surfaces to be cut up into separate components that are handled individually. Given appropriate boundary conditions, it can compute a unique circular slit map up to a rotation around the center. The slit mapping computes the intrinsic structure of the given surface, which can be reflected in the shape of the target domain. Then the resultant parameterization is used for surface registration.
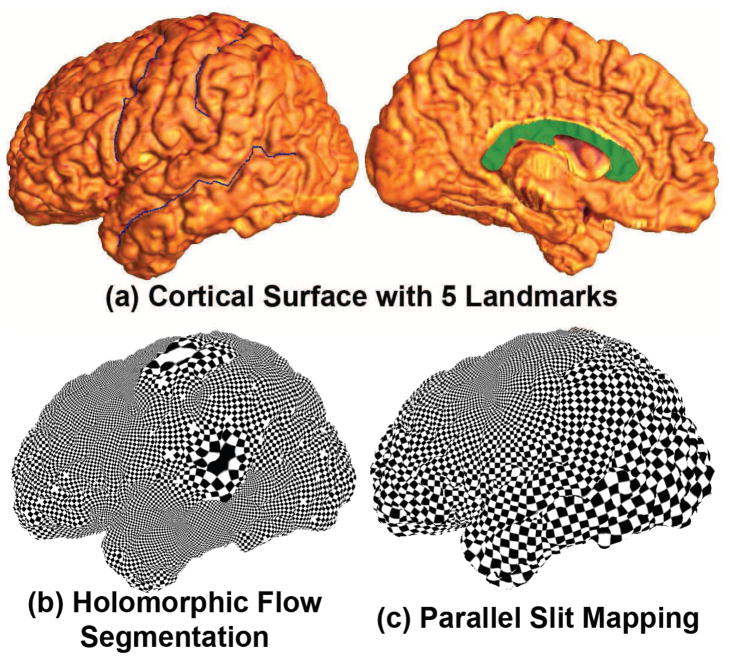
In our work, by cutting along specific landmark curves lying on the cortical surfaces, we convert a cortical surface into a multiply connected surface. We call this topology optimization. With topology optimization, we turn a limited number of important geometric features into boundaries and match them by enforcing the boundary matching conditions (Wang et al., 2008b) or analyze geometric feature statistics around these boundaries (Wang et al., 2009b). Three conformal parameterization methods, the holomorphic flow segmentation method (Wang et al., 2007), the surface Ricci flow method (Wang et al., 2006; Wang et al., 2012b) and the slit mapping method, can parameterize brain cortical surfaces with boundaries, i.e. multiply connected surfaces. Fig. 13 compares two conformal parameterization methods, the holomorphic flow method (Wang et al., 2007) and the parallel slit mapping method. To compute the conformal parameterization, the holomorphic flow method needs to use a “double covering” to turn the surface into a closed surface. The resulting conformal parameterization has singularities which cause big distortions around the singularity areas. Slit mapping works on open boundary surfaces, so that the computational cost is lower. The result does not contain singularities and we also have the flexibility to assign which of two boundaries comprise the outer and inner circles for the circular slit mapping or two outer boundaries for parallel slit mapping. Fig. 13 (a) shows a left hemisphere cortical surface with 5 landmarks: the Precentral Sulcus, Postcentral Sulcus, Superior Temporal Sulcus (Main Body), and linked Callosal Sulcus and Inferior Callosal Outline Segment. The resulting conformal parameterizations are shown by texture mapping of a checkerboard onto the surface. Fig. 13 (b) shows the parameterization result based on holomorphic flow (Wang et al., 2007) and (c) is the parallel slit map. In both pictures, the right angles are preserved but there are two zero points on the holomorphic flow result. However, there is no singularity on the parallel slit mapping result and the parameterization result is relatively uniform over the whole surface. This uniformity is likely to be beneficial for accurate assessment of brain morphometry.
Figure 13.
Comparison of slit mapping to holomorphic flow segmentation, as a method to parameterize the cortical surface. The first row shows a left cortical surface with 5 selected landmark curves. The second row shows results of holomorphic flow segmentation (Wang et al., 2007) and parallel slit mapping. Both algorithms work with surfaces that have boundaries. However the holomorphic flow segmentation method generates singularities in the parameterization results (large tiles in (b)).
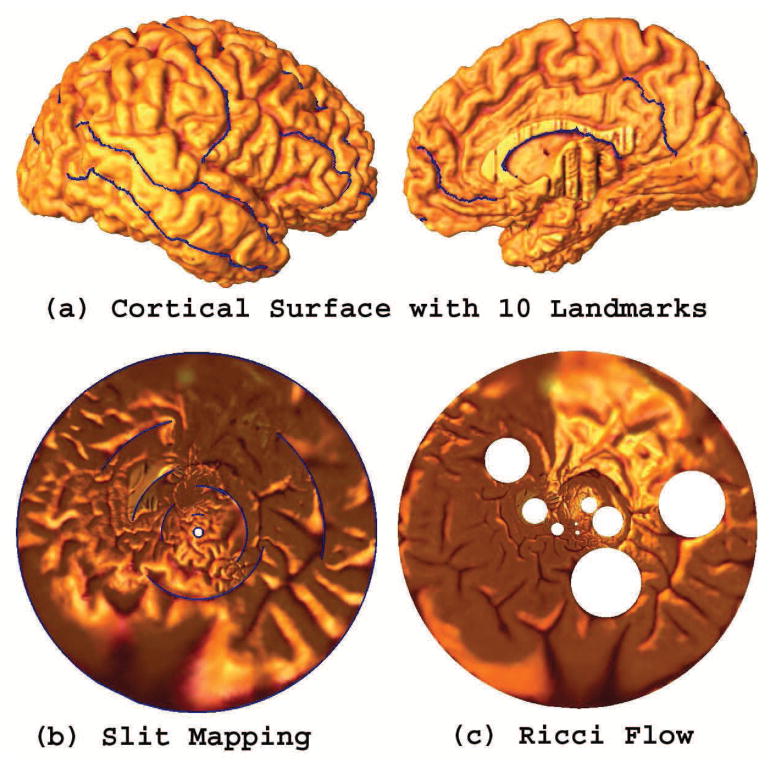
Fig. 14 compares the surface Ricci flow method (Wang et al., 2006; Wang et al., 2012b) and the circular slit mapping method, on a right cerebral cortical surface with 10 landmarks as described in Sec. 3. After cutting along these landmark curves, the cerebral cortical surface is turned into an open boundary surface with 10 boundaries. Ricci flow can also directly work on the multiply connected surface. However, it is a nonlinear optimization process, which is more time-consuming than slit mapping. Slit mapping involves solving a linear system so the computation is easier and stable. The surface with 10 landmarks overlaid is shown in Fig. 14 (a). The respective parameterization results are shown in (b) (circular slit map) and (c) (Ricci flow). Neither result has any singularities, and both of them offer a canonical space for cortical surface registration (Wang et al., 2008b). However, on the unit disk, the single empty hole in the circular slit map occupies 0.07% of the unit disk area while the 9 empty holes in the surface Ricci flow method result occupy 10.46% of the unit disk area. When we use the unit disk for surface parameterization and registration, intuitively, a parameter space without holes is better. When comparing these two unit disk parameterizations for surface mapping, the circular slit mapping result may be more practicable than using surface Ricci flow.
Figure 14.
Comparison of slit mapping with the Ricci flow, as methods to parameterize the cortical surface. The first row shows a right hemisphere cortical surface, with 10 selected landmark curves. The second row shows results of using the circular slit mapping and Ricci flow methods (Wang et al., 2006). Both algorithms work with a surface that has boundaries. However the Ricci flow is computed by solving a nonlinear system, so it is less efficient than the slit map conformal parameterization method.
Stability of Conformal Maps in Longitudinal Scans
The proposed slit map method is based on solving a linear system of differential forms. The differential forms can be easily represented as a vector associated with each edge in our current surface representation. So the method is stable and can be readily extended to a surface with multiple boundaries.
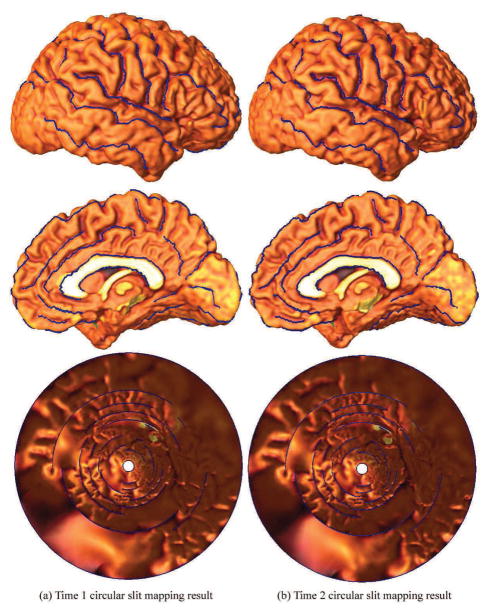
Fig. 15 shows (visually at least) that the parameterization is consistent on a pair of right hemisphere cortical surfaces from the same healthy control subject (Thompson et al., 2003) scanned twice, with a 3.1 years interval. Most of the surface shape was stable with slight differences due to development. In prior work (Thompson et al., 2003), 35 landmark curves were labeled on each cortical surface as detailed in (Sowell et al., 2002) and (Hayashi et al., 2002). The cortical surfaces are shown in Fig. 15, with landmark curves in blue. As an ongoing effort, we are studying the stability of the slit mapping method using different numbers of available landmark curves. Computational results are shown on the last row in Fig. 15. Each cortical surface is conformally mapped to an annulus with 33 concentric slits. We can see that the positions and lengths of the slits are very similar in the two situations. Although more data and computations are clearly necessary, this illustration suggests feasibility and stability of using our conformal slit mapping method, with different number of landmarks.
Figure 15.
Slit mapping on longitudinal cortical surfaces, from the same subject scanned twice. A right hemisphere cortical surface extracted from the same control subject at two different times in our prior study (Thompson et al., 2003). All available landmarks (35 in total) are used as cuts in the surface, allowing the entire surface to be mapped to a unit disk with 33 inner slits and 1 inner circle. The two parameterization results are very similar. Although more empirical data would be useful, this suggests the robustness of our proposed method if a person is re-scanned.
Sulcal landmark curve selection
In our dataset, there are 36 sulcal landmarks on each hemisphere cortical surface and a total of 72 sulcal landmark curves on the whole brain (Hayashi et al., 2002; Sowell et al., 2002). When we initially defined these protocols, 10 years ago, we labeled every possible sulcus from the Ono atlas that could be reliably defined on the brain surfaces of multiple people. A committee of several neurologists and anatomists came up with a protocol that would be feasible for trained anatomists to use reliably, with good inter-rater agreement, even for tracers who had worked independently. In the end, because the protocol was so time consuming to perform, not many investigators were willing to hand-landmark so many curves, even though we did so for this dataset. This motivated us to reduce the total number of sulcal curves used in the registration.
Different studies have historically used different choices of landmark curves. For example, in Pantazis et al. (2010), a set of 26 landmarks was defined per hemisphere while 28 landmark curves were used in Joshi et al. (2012b) and only 14 sulcal curves were used in Zhong and Qiu (2010). In this work, among 36 sulcal landmark curves per hemisphere, we used 10 landmark curves: the Central Sulcus, Superior Temporal Sulcus (Main Body), Inferior Frontal Sulcus, Middle Frontal Sulcus, Inferior Temporal Sulcus, Secondary Intermediate Sulcus, Transverse Occipital Sulcus, Inferior Callosal Outline Segment, Superior Rostral Sulcus, and Subparietal Sulcus. This set of landmarks includes many of the major features of the brain, including landmarks that are scattered across all lobes, especially including the longer sulci. Our experiments on both group difference and classification demonstrated reasonable effectiveness with this selected landmark set. We agree that the current empirical study does not prove that it is the best landmark subset for disease classification, and it would certainly be interesting in the future to optimize the chosen set of landmarks with respect to the classification performance. In the end, one would imagine that classification performance would increase with the number of landmarks chosen, so long as the additional landmarks were in areas that show differences between the groups being classified.
Two-stage Lasso
In our classification experiments, we employed a two-stage approach where Lasso is used to select features and a subsequent ridge regression for the classification. Lasso employs a convex regularization term (ℓ1-norm) to approximate the combinatorial ℓ0-norm to make the problem computationally tractable. However, Lasso is known to induce estimation bias due to the convex relaxation; Lasso not only shrinks the coefficients of the irrelevant variables to zero (these features are removed), but also shrinks the coefficients of the relevant variables. In the literature, many methods have been proposed to correct the unnecessary shrinkage, e.g., two-stage Lasso and multi-stage Lasso (Zou, 2006; Zhang, 2010). Since Lasso is reasonably good at identifying relevant features, it is natural to apply a two-stage approach like ours, in which the features are identified at the first stage and the coefficients are learnt at the second stage to reduce the shrinkage effect. It is also common to add a small ℓ2-norm to the Lasso formulation, which follows the elastic nets formulation (Zou and Hastie, 2005) and overcomes the limitations of the Lasso method. For example, in our SLEP package (Liu et al., 2009), we add a ℓ2-norm regularization to the Lasso formulation. Our experience showed that it generally improved Lasso performance (Ye et al., 2012).
Leave-N-out Experiments
Besides the leave-one-out experiments, we also performed leave-N-out experiments to help illustrate the mean classification accuracy and its standard deviation when varying the training and testing datasets by random re-sampling. As our current dataset had only 82 subjects, we initially focused on leave-one-out experiments, to maximize the size of the training set. For completeness, we also performed leave-N-out experiments, with mTBM and TBM features, to see how the mean accuracy in a larger test set depended on the test set chosen. In our experiments, for each round, we randomly selected 10 out of 82 images for testing and used the remaining 72 images for training. During training, a cross-validation scheme was adopted to set up the parameter values. The selected parameter values were then applied to test the classifier on the testing data. We further repeated this process 30 times. For each time, an accuracy rate was calculated on the testing dataset. After all rounds of experiments, we obtained a set of accuracy rates from the leave-N-out experiments. We further estimated the mean accuracy rates and their standard deviations. We note that the standard deviation here measures the accuracy on repeated random drawings of 10 subjects from the sample, which were left out from the training set to guarantee independence and avoid circularity. We performed experiments using both TBM and mTBM algorithms and their experimental results (mean accuracies and standard deviations) are summarized in Table 9.
Table 9.
Mean accuracy and standard deviation results for leave-N-out experiments with mTBM and TBM. Results are shown for two-sample t-tests that test whether the difference between the two average accuracy rates is statistically different. These results are shown as p-values in the last column of the table. Interestingly, the multivariate TBM method does not outperform the standard TBM method applied to surfaces.
| mTBM | TBM | p-value | |
|---|---|---|---|
| Left hemisphere | 0.66 ± 0.150 | 0.68 ± 0.161 | 0.62 |
| Right hemisphere | 0.76 ± 0.113 | 0.77 ± 0.123 | 0.66 |
| Whole brain | 0.68 ± 0.130 | 0.65 ± 0.125 | 0.32 |
In line with the leave-one-out experiments – right hemisphere features tended to perform better in discriminating the two classes (2 sample t-test, right vs. left, p=0.0051; right vs. whole, p=0.0136). As seen from the standard deviations, the classification accuracy rate did not have a wide range. We computed 30 accuracy rates using different training/testing partitions, but none of the comparisons was significant. Surprisingly perhaps, the multivariate TBM method did not outperform the standard method. This may be due to the relatively small size of the current dataset. It could be that the benefit of learning of the multivariate parameters in TBM is either minimal, or requires a very large sample size to detect.
Group Difference Performance vs. Classification Performance
Overall, these leave-one-out experiments correctly classified 67/82 cases for TBM and 65/82 cases for mTBM. A binomial proportion test can be performed on these two proportions, with the null hypothesis that these two accuracy rates are the same. The p-value from this test was 0.6935, which shows that no one method had statistically superior accuracy. While mTBM achieved the strongest group difference, when compared to three other TBM-based and volume features, it achieved classification accuracy comparable to univariate TBM.
Based on the results, we have demonstrated that MRI-based disease classification can benefit from surface parameterization with differential forms and tensor-based morphometry, in the log-Euclidean domain, on the resulting surface tensors. The similar classification performance of mTBM and TBM may be due to the limited sample size for the dataset used in our current experiments, although we know that 82 subjects is an adequately powered dataset to detect group differences, as we were able to detect them. It could be that multivariate surface descriptors require a larger sample size to show benefits over the scalar descriptors in classification accuracy, to more accurately learn their covariance structure. Or, it may be that multivariate TBM can outperform scalar TBM in some contexts but not others, or in some diseases but not others, depending on the type of surface differences there are. A strong group difference result does not always guarantee a better classification power; this is consistent with discoveries reported in some prior work (Lao et al., 2004; Chincarini et al., 2011; Cuingnet et al., 2011). Our longer-term goal is to investigate how to best apply surface tensor-based morphometry (Davatzikos et al., 1996; Thompson et al., 2000a; Woods, 2003; Chung et al., 2008; Wang et al., 2010c) in disease classification research.
Improvement of classification performance with spatial and anatomical regularization
Our current work proposed a surface-based framework which demonstrated that, by analyzing the full surface deformation tensors computed from a set of parametric surfaces, one may achieve improved group difference detection and group classification performance. In our experiments, with the ℓ1-norm penalty (sparse learning) methods, we achieved reasonable classification results. However, there is a potential to further improve classification performance by considering spatial and anatomical regularization in medical imaging research. Pioneering work has demonstrated that spatial and anatomical information can enhance performance of typical classifiers, such as support vector machine (SVM) (Fung and Stoeckel, 2007; Cuingnet et al., 2010), Adaboost (Xiang et al., 2009) and Bayesian classifier (Sabuncu and Van Leemput, 2011), improve feature reduction efficiency (Batmanghelich et al., 2012) and behavior prediction accuracy (Michel et al., 2011). The key insight is that the underlying anatomical structures are smooth, i.e. features in biomedical applications exhibit certain intrinsic structures, such as groups, graphs, and trees (Jacob et al., 2009; Li and Li, 2010; Liu and Ye, 2010b; Liu et al., 2010a; Liu and Ye, 2010a); for example, if a surface point shows significant difference between two groups, most likely its neighboring surface points also have certain statistical discrimination power. More importantly, a single surface point may not carry strong statistical power of its own, but a set of such correlated points may do so. In other words, an association mapping approach may “borrow strength” from correlated phenotypes and can potentially yield higher statistical power (Ferreira and Purcell, 2009). Recently, we and other groups have developed structured sparse learning formulations which incorporate the complex feature structures into the sparse learning model; they have been applied successfully in many applications including computer vision, medical imaging and bioinformatics (Zou and Hastie, 2005; Yuan and Lin, 2006; Jacob et al., 2009; Zhao et al., 2009; Kim and Xing, 2010; Li and Li, 2010; Liu and Ye, 2010b; Liu et al., 2010a; Liu and Ye, 2010a; Wang et al., 2011a; Wang et al., 2012a). In future, we plan to develop advanced structured sparse learning strategies for surface-based disease marker identification with the potential of leading to major discoveries in neuroimaging.
Alternatives for full deformation tensor information processing
In our work, we analyzed full deformation tensor information by using log-Euclidean framework. However, there are other ways available to analyze the full tensor information. A simple strategy is to directly use a Euclidean structure on square matrices to define a metric on the tensor space. But the classical Euclidean framework has many defects, for example, null or negative eigenvalues appear during Euclidean computation. To fully circumvent these difficulties, affine-invariant Riemannian metrics have been proposed for tensors (Fletcher and Joshi, 2004; Moakher, 2005; Pennec et al., 2006) and a class of tensor interpolation paths, geodesic-loxodrome (Kindlmann et al., 2007), was proposed to explicitly preserve clinically important tensor attributes. While the log-Euclidean framework is the most computationally convenient, other approaches (Fletcher and Joshi, 2004; Moakher, 2005; Pennec et al., 2006; Kindlmann et al., 2007) also provide valid, theoretically rigorous alternatives for full anatomical deformation tensor analysis.
Beyond the current system
In this paper, we validated our proposed work in a dataset used in our prior work (Thompson et al., 2005). In Thompson et al. (2005), an operator who was blind to the subject’s pathological condition has normalized identified certain landmark points. In this scenario, when a new subject was given to the system, the operator identified a set of landmark points using the International Consortium for Brain Mapping-53 average brain (Mazziotta et al., 2001; Thompson et al., 2005). The identified landmark points were used to compute a rigid-body transformation that aligns brain volume images in a standard space for segmentation and cortical surface generation. The generated surfaces and 10 manually labeled landmark curves on the surfaces were the input to our method to apply multivariate statistics for analyzing the group difference and classification. However, our method is general enough to process landmark curves that are either manually traced or automatically detected by some other software tools. We are exploring the possibility to take the landmark curves detected by BrainSuite (Joshi et al., 2012a) as input for conformal slit map based surface registration. Our ongoing work (An et al., 2012) also tries to apply multivariate tensor-based morphometry on cortical surface built by FreeSurfer (Fischl et al., 1999).
5. CONCLUSION
We presented an MRI-based computer-assisted diagnostic classification system that finds vertices and local features on cortical surface models to best discriminate two groups of subjects. Our system was based on mTBM and sparse learning, with the ℓ1-norm based penalty. The mTBM captured the full deformation tensor information and performed better than TBM (the determinant of Jacobian matrix) in both group difference and classification studies. The sparse learning method selected a smaller, biologically plausible feature set, consistent with prior knowledge of cortical anomalies features in WS. In leave-one-out tests, with the same parameter settings, our surface-based method achieved better classification results than those using the pair of eigenvalues of Jacobian matrix and the whole cortical volumes as features. Ongoing work is testing this framework for studying structural imaging biomarkers of other disorders, such as Alzheimer’s disease.
Table 6.
Classification performance with pair of eigenvalues of Jacobian matrix on right cortical hemisphere, left cortical hemisphere and both cortical hemispheres.
| Pair EV of J on right cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 22 | 20 | |
| CTL | 3 | 37 | |
| Pair EV of J on left cortical hemisphere | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 21 | 21 | |
| CTL | 5 | 35 | |
| Pair EV of J on both cortical hemispheres | True Class | Assigned Class | |
| WS | CTL | ||
| WS | 18 | 24 | |
| CTL | 4 | 36 | |
APPENDIX
The following sections describe the mathematics underlying the slit mapping algorithm and how we measure features on the cortical surfaces. Some concepts come from exterior calculus and use the terminology of differential forms, which is useful for analyzing surfaces represented as parametric meshes.
A.1. Theoretic background on holomorphic 1-forms
Suppose S is a surface embedded in ℝ3, with induced Riemannian metric g. S is covered by an atlas {(U, φ)}. Let (x, y) be the local parameter on the chart (U, φ). If the metric g has the representation g = e2λ(x,y) (dx2 + dy2), we say (x, y) is isothermal. An atlas consisting of isothermal parameter charts is called a conformal structure.
The Laplace-Beltrami operator on a surface is defined as
| (6) |
A function defined on a surface f : S → ℝ is harmonic, if Δgf ≡ 0.
Mathematically, the differential form is a concept used in multivariable calculus that is independent of any chosen set of coordinates - it has many applications in geometry, topology and physics. The general setting for the study of differential forms is on a differential manifold. A differential 1-form in the local parameters (x, y) may be defined as
| (7) |
where f, g are smooth functions. Differential 1-forms are naturally dual to vector fields on a manifold. The algebra of differential forms along with the exterior derivative defined on it is preserved under smooth functions between two manifolds. Specifically, ω is a closed 1-form, if in each local parameter (x, y), . If ω is the gradient of another function defined on S, it can be called an exact 1-form. An exact 1-form is also a closed 1-form. If a closed 1-form ω satisfies , then it is a harmonic 1-form. The gradient of a harmonic function is an exact harmonic 1-form.
The Hodge star operator acting on a differential 1-form gives the conjugate differential 1-form
| (8) |
Intuitively, the conjugate 1-form *ω is obtained by rotating ω by a right angle everywhere. If ω is harmonic, so is its conjugate *ω.
Built upon the harmonic 1-form concept, the holomorphic 1-form is a differential form on a manifold which can have complex coefficients. It consists of a pair of conjugate harmonic 1-forms
| (9) |
If two harmonic fields are orthogonal everywhere, they form a holomorphic 1-form. An intrinsic way to compute conformal parameterization is to search for a holomorphic 1-form that satisfies certain properties. In conformal slit mapping, we shall find certain holomorphic 1-forms with special behavior on the boundaries of the surface.
Suppose S is an open surface with n boundaries γ1, γ2, … γn, we can uniquely find a holomorphic 1-form τ, such that (Ahlfors, 1953)
| (10) |
Definition 1 (Circular Slit Mapping)
Fix a point p0 on the surface S, for any point p ∈ S, let γ be an arbitrary path connecting p0 and p, then the circular slit mapping is defined as φ(p) = e∫γτ.
Theorem 1
The function φ effects a one-to-one conformal mapping of surface S onto the annulus 1 < |z| < eλ0−n − 2 concentric arcs situated on the circles |z| = eλi, i = 1,2, … n − 2.
The proof of the above theorem on slit mapping may be found in (Ahlfors, 1953). For a given choice of the inner and outer circle, the circular slit mapping is uniquely determined up to a rotation around the center. The parallel slit mapping can be defined in a similar way.
Definition 2 (Parallel Slit Mapping)
Let S̄ be the universal covering space of the surface S, π : S̄ → S be the projection and τ̄ = π*τ be the pull back of τ. Fix a point p̄0 on S̄, for any point p̄ ∈ S, let γ̄ be an arbitrary path connecting p̄0 and p̄, then the parallel slit mapping is defined as φ̄(p̄) = ∫γ̄τ̄.
In our work, we employ the concepts of holomorphic 1-form and conformal slit mapping to compute a conformal parameterization of cortical surfaces. On the Euclidean parameter domain, the constrained harmonic map is computed to construct correspondences between cortical surfaces.
A.2. Computation of Surface Conformal Slit Mapping
In engineering fields, smooth surfaces are often approximated by simplicial complexes (triangle meshes). Major concepts in the continuous setting can be generalized to the discrete setting. In this section, we briefly describe the algorithm implementation to compute the 1-forms and slit mapping.
Suppose M is a triangle mesh with n +1 boundaries, denoted as ∂M = γ0 – γ1 –··· – γn. We use vi to denote a vertex, which is also a 3D coordinate vector, [vi, vj] denote an edge connecting vertices vi and vj, [vi, vj, vk] denote a triangle formed by vertices vi, vj, and vk. The angle at vertex vi in triangle [vi, vj, vk] is denoted as . The discrete functions defined on vertices, edges, and faces are called discrete 0-forms, 1-forms, and 2-forms, respectively. The discrete divergence, discrete Laplace-Beltrami operator and discrete wedge operator can be defined as reported in our prior work (Wang et al., 2008a; Wang et al., 2010c).
The algorithm pipeline is as follows:
Compute the basis for all exact harmonic 1-forms;
Compute the basis for all harmonic 1-forms;
Compute the basis for all holomorphic 1-forms;
Construct the slit mapping.
The detailed algorithms to compute 1-forms are reported in our earlier work (Wang et al., 2008a; Wang et al., 2010c). Here we give a brief description of the pipeline implementation. Without loss of generality, we map the boundary γ0 to the outer circle of the circular slit domain, γ1 to the inner circle and all other boundaries to the concentric slits. Briefly, first, for each inner boundary γ1, … γn, a harmonic function is computed by solving a Dirichlet problem (Wang et al., 2008a; Wang et al., 2010c). By definition, the exact harmonic 1-form ωk can be computed as the gradient of the harmonic function on boundary γk, (k = 1, 2, …, n). We denote the exact harmonic 1-forms basis as {ω1, ω2, …, ωn}. Second, we compute the closed but non-exact harmonic 1-form τk along the path connecting the inner boundary γk, (k = 1, 2, … n) to outer boundary γ0 (Wang et al., 2008a; Wang et al., 2010c). The closed harmonic 1-form basis is the union of the exact harmonic 1-forms basis {ω1, ω2, …, ωn} and the closed but non-exact harmonic 1-forms basis {τ1, τ2, …, τn}. The computed closed harmonic 1-forms basis is used to improve the computational accuracy of the conjugate 1-form *ωk of the exact harmonic 1-form ωk because of the inaccuracy of directly applying the brute-force Hodge star operator (Eq. 8). We denote the conjugate 1-forms basis as {*ω1, *ω2, …, *ωn} (Wang et al., 2008a; Wang et al., 2010c). Then, the holomorphic 1-forms basis is { } (Eq. 9). Next, for surface conformal slit mapping, we need to find a special holomorphic 1-form , such that the imaginary part of its integral satisfies
| (11) |
To get the coefficients λi, we solve the following linear system for λi, i = 1,2, …, n:
| (12) |
where αjk = ∫γj*ωk.7
It can be proved that this linear system has a unique solution, which reflects the fact that γ1 is mapped to the inner circle of the circular slit domain. Further, the system implies λ1α01 + λ2α02 + … + λnα0n = 2π, which means that γ0 is mapped to the outer circle in the circular slit domain. The circular slit mapping is a complex-valued function φ : M → ℂ. Choosing a base vertex v0 arbitrarily, and for each vertex v ∈ M choosing the shortest path γ from v0 to v, the map can be computed as φ(v) = e∫γω. Based on the circular slit map φ, we can compute a parallel slit map φ : M → ℂ as φ(v) = lnφ(v).
Footnotes
Note 72 landmark curves, as defined in (Sowell et al., 2003), were originally traced in (Thompson et al. 2005) but we only used 20 of them here.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlfors LV. Complex Analysis. McGraw-Hill; New York: 1953. [Google Scholar]
- An X, Shi J, Wang Y. Cortical Differences Analysis with Multivariate Tensor-Based Morphometry in 829 ADNI subjects. The 18th Annual meeting of the Organization for Human Brain Mapping; Beijing, China. 2012. [Google Scholar]
- Angenent S, Haker S, Tannenbaum A, Kikinis R. On the Laplace-Beltrami operator and brain surface flattening. IEEE Trans Med Imaging. 1999;18(8):700–711. doi: 10.1109/42.796283. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, Mistur R, Tsui WH, de Leon MJ. Subregional hippocampal atrophy predicts Alzheimer’s dementia in the cognitively normal. Neurobiol Aging. 2010a;31(7):1077–1088. doi: 10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM, Green AE, Hwang KS, Zoumalan C, Jack CR, Jr, Harvey DJ, Petersen RC, Thal LJ, Aisen PS, Toga AW, Cummings JL, DeCarli CS. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp. 2010b;31(5):786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean Metrics for Fast and Simple Calculus on Diffusion Tensors. Magn Reson Med. 2006;56(2):411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: deformation-based morphometry. Human Brain Mapping. 1998;6(5–6):348–357. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Auzias G, Glaunès J, Colliot O, Perrot M, Mangin J-F, Trouvé A, Baillet S. DISCO: A Coherent Diffeomorphic Framework for Brain Registration under Exhaustive Sulcal Constraints. Med Image Comp Comput-Assist Intervention, Proceedings. 2009:730–738. doi: 10.1007/978-3-642-04268-3_90. [DOI] [PubMed] [Google Scholar]
- Bakircioglu M, Joshi S, Miller MI. Landmark Matching on Brain Surfaces via Large Deformation Diffeomorphisms on the Sphere. Proc SPIE Medical Imaging. 1999:710–715. [Google Scholar]
- Bansal R, Staib LH, Xu D, Zhu H, Peterson BS. Statistical analyses of brain surfaces using Gaussian random fields on 2-D manifolds. IEEE Trans Med Imaging. 2007;26(1):46–57. doi: 10.1109/TMI.2006.884187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batmanghelich NK, Taskar B, Davatzikos C. Generative-discriminative basis learning for medical imaging. IEEE Trans Med Imaging. 2012;31(1):51–69. doi: 10.1109/TMI.2011.2162961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A, Teboulle M. A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM Journal on Imaging Sciences. 2009;2(1):183–202. [Google Scholar]
- Bruckstein AM, Donoho DL, Elad M. From Sparse Solutions of Systems of Equations to Sparse Modeling of Signals and Images. SIAM Review. 2009;5(1):34–81. [Google Scholar]
- Brun C, Lepore N, Pennec X, Chou YY, Lee AD, Barysheva M, de Zubicaray G, Meredith M, McMahon K, Wright MJ, Toga AW, Thompson PM. A tensor-based morphometry study of genetic influences on brain structure using a new fluid registration method. Med Image Comput Comput Assist Interv. 2008;11(Pt 2):914–921. doi: 10.1007/978-3-540-85990-1_110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. Feature-based fusion of medical imaging data. IEEE Trans Inf Technol Biomed. 2009;13(5):711–720. doi: 10.1109/TITB.2008.923773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candès EJ, Tao T. Decoding by linear programming. Information Theory, IEEE Transactions on. 2005;51(12):4203–4215. [Google Scholar]
- Candès EJ, Romberg J, Tao T. Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. Information Theory, IEEE Transactions on. 2006;52(2):489–509. [Google Scholar]
- Candès EJ, Wakin M. An introduction to compressive sampling. IEEE Signal Processing Magazine. 2008;25(2):21–30. [Google Scholar]
- Cao J, Worsley KJ. The detection of local shape changes via the geometry of Hotelling’s T2 fields. Ann. Statist. 1999;27(3):925–942. [Google Scholar]
- Carmichael OT, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Kuller LH, Lopez OL, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Mapping ventricular changes related to dementia and mild cognitive impairment in a large community-based cohort. Biomedical Imaging: Nano to Macro, 2006; 3rd IEEE International Symposium on; 2006. pp. 315–318. [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Acceleration of cerebral ventricular expansion in the Cardiovascular Health Study. Neurobiology of Aging. 2007a;28(1):1316– 1321. doi: 10.1016/j.neurobiolaging.2006.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Cerebral Ventricular Changes Associated With Transitions Between Normal Cognitive Function, Mild Cognitive Impairment, and Dementia. Alzheimer’s Disease and Associated Disorders. 2007b;21(1):14– 24. doi: 10.1097/WAD.0b013e318032d2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Ventricular volume and dementia progression in the Cardiovascular Health Study. Neurobiology of Aging. 2007c;28(3):389–397. doi: 10.1016/j.neurobiolaging.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Huan Z, Caselli RJ, Bandy D, Ayutyanont N, Alexander GE. Linking functional and structural brain images with multivariate network analyses: a novel application of the partial least square method. NeuroImage. 2009;47(2):602–610. doi: 10.1016/j.neuroimage.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Ayutyanont N, Langbaum JBS, Fleisher AS, Reschke C, Lee W, Liu X, Bandy D, Alexander GE, Thompson PM, Shaw L, Trojanowski JQ, Jack CR, Jr, Landau SM, Foster NL, Harvey DJ, Weiner MW, Koeppe RA, Jagust WJ, Reiman EM. Characterizing Alzheimer’s disease using a hypometabolic convergence index. Neuroimage. 2011;56(1):52–60. doi: 10.1016/j.neuroimage.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, Rei L, Squarcia S, Rodriguez G, Bellotti R, Cerello P, De Mitri I, Retico A, Nobili F. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage. 2011;58(2):469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Chou Y, Leporé N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. NeuroImage. 2008;40(2):615– 630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y, Leporé N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. NeuroImage. 2009;46(2):394– 410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Robbins S, Paus T, Taylor J, Giedd JN, Rapoport JL, Evans AC. Deformation-based Surface Morphometry Applied to Gray Matter Deformation. NeuroImage. 2003;18:198–213. doi: 10.1016/s1053-8119(02)00017-4. [DOI] [PubMed] [Google Scholar]
- Chung MK, Dalton KM, Davidson RJ. Tensor-Based Cortical Surface Morphometry via Weighted Spherical Harmonic Representation. IEEE Trans Med Imag. 2008;27(8):1143–1151. doi: 10.1109/TMI.2008.918338. [DOI] [PubMed] [Google Scholar]
- Combettes PL, Pesquet JC. Arxiv. 2009. Proximal splitting methods in signal processing. preprint arXiv:0912.3522. [Google Scholar]
- Correa NM, Adali T, Li YO, Calhoun VD. Canonical Correlation Analysis for Data Fusion and Group Inferences: Examining applications of medical imaging data. IEEE Signal Process Mag. 2010;27(4):39–50. doi: 10.1109/MSP.2010.936725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu CY, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Wahlund LO, Spenger C, Toga AW, Lovestone S, Simmons A. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage. 2011;56(1):212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuingnet R, Chupin M, Benali H, Colliot O. Spatial and anatomical regularization of SVM for brain image analysis. In: Lafferty J, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, editors. Advances in Neural Information Processing Systems. Vol. 23. 2010. pp. 1–9. [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage. 2011;56(2) doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Spatial Normalization of 3D Brain Images using Deformable Models. J Comp Assisted Tomography. 1996;20(4):656–665. doi: 10.1097/00004728-199607000-00031. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Vaillant M, Resnick SM, Prince JL, Letovsky S, Bryan RN. A computerized approach for morphological analysis of the corpus callosum. J Comput Assist Tomogr. 1996;20(1):88–97. doi: 10.1097/00004728-199601000-00017. [DOI] [PubMed] [Google Scholar]
- Donoho DL. Compressed sensing. Information Theory, IEEE Transactions on. 2006;52(4):1289–1306. [Google Scholar]
- Drury HA, Van Essen DC, Anderson CH, Lee CW, Coogan TA, Lewis JW. Computerized Mappings of the Cerebral Cortex: A Multiresolution Flattening Method and a Surface-based Coordinate System. J Cognitive Neurosciences. 1996;8:1–28. doi: 10.1162/jocn.1996.8.1.1. [DOI] [PubMed] [Google Scholar]
- Durrleman S, Pennec X, Trouvé A, Thompson P, Ayache N. Inferring brain variability from diffeomorphic deformations of currents: An integrative approach. Medical Image Analysis. 2008;12(5):626–637. doi: 10.1016/j.media.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen D, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Med Image Comput Comput Assist Interv. 2005;8(Pt 1):1–8. doi: 10.1007/11566465_1. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. COMPARE: classification of morphological patterns using adaptive regional elements. IEEE Trans Med Imaging. 2007;26(1):93–105. doi: 10.1109/TMI.2006.886812. [DOI] [PubMed] [Google Scholar]
- Ferrarini L, Palm WM, Olofsen H, van der Landen R, van Buchem MA, Reiber JHC, Admiraal-Behloul F. Ventricular shape biomarkers for Alzheimer’s disease in clinical MR images. Magnetic resonance in medicine. 2008;59(2):260–267. doi: 10.1002/mrm.21471. [DOI] [PubMed] [Google Scholar]
- Ferreira MAR, Purcell SM. A multivariate test of association. Bioinformatics. 2009;25(1):132–133. doi: 10.1093/bioinformatics/btn563. [DOI] [PubMed] [Google Scholar]
- Figueiredo MAT, Nowak RD, Wright SJ. Gradient projection for sparse reconstruction: Application to compressed sensing and other inverse problems. Selected Topics in Signal Processing, IEEE Journal of. 2007;1(4):586–597. [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical Surface-Based Analysis II: Inflation, Flattening, and a Surface-Based Coordinate System. NeuroImage. 1999;9(2):195– 207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fletcher T, Joshi S. In: Jan, editor. Principal Geodesic Analysis on Symmetric Spaces: Statistics of Diffusion Tensors; Computer Vision and Mathematical Methods in Medical and Biomedical Image Analysis: ECCV 2004 Workshops CVAMIA and MMBIA Prague; Czech Republic. May 15, 2004; Springer; 2004. Revised Selected Papers. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung G, Stoeckel J. SVM feature selection for classification of SPECT images of Alzheimer’s disease using spatial information. Knowl Inf Syst. 2007;11(2):243–258. [Google Scholar]
- Golland P, Grimson WEL, Shenton ME, Kikinis R. Deformation Analysis for Shape Based Classification. Proceedings of the 17th International Conference on Information Processing in Medical Imaging; Springer-Verlag. 2001. pp. 517–530. [Google Scholar]
- Groves AR, Beckmann CF, Smith SM, Woolrich MW. Linked independent component analysis for multimodal data fusion. Neuroimage. 2011;54(3):2198–2217. doi: 10.1016/j.neuroimage.2010.09.073. [DOI] [PubMed] [Google Scholar]
- Gu X, Wang Y, Chan TF, Thompson PM, Yau S-T. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Trans Med Imag. 2004;23(8):949–958. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- Gutman B, Wang Y, Morra J, Toga AW, Thompson PM. Disease classification with hippocampal shape invariants. Hippocampus. 2009;19(6):572–578. doi: 10.1002/hipo.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene Selection for Cancer Classification using Support Vector Machines. Mach Learn. 2002;46(1–3):389–422. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; New York, NY: 2001. [Google Scholar]
- Hayashi KM, Thompson PM, Mega MS, Zoumalan CI, Dittmer SS. Medial hemispheric surface gyral pattern delineation in 3D: surface curve protocol. 2002 Available from: http://www.loni.ucla.edu/~khayashi/Public/medial_surface/
- Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multilocus genetic analysis of brain images. Front Genet. 2011;2:73. doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. The generalization of Student’s ratio. Ann Math Statist. 1931;2:360–378. [Google Scholar]
- Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, Leow AD, Toga AW, Jack CR, Jr, Harvey D, Weiner MW, Thompson PM. Unbiased tensor-based morphometry: Improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, Chen K, Reiman E. Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. NeuroImage. 2010;50(3):935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurdal MK, Stephenson K. Cortical cartography using the discrete conformal approach of circle packings. Neuroimage. 2004;23(Suppl 1):S119–128. doi: 10.1016/j.neuroimage.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Hurdal MK, Stephenson K. Discrete conformal methods for cortical brain flattening. Neuroimage. 2009;45(1 Suppl):S86–98. doi: 10.1016/j.neuroimage.2008.10.045. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, Bernstein MA, Gunter JL, Pankratz VS, Aisen PS, Weiner MW, Petersen RC, Shaw LM, Trojanowski JQ, Knopman DS. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133(11):3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob L, Obozinski G, Vert J-P. Group lasso with overlap and graph lasso. Proceedings of the 26th Annual International Conference on Machine Learning; ACM, Montreal, Quebec, Canada. 2009. pp. 433–440. [Google Scholar]
- Ji S, Yuan L, Li Y-X, Zhou Z-H, Kumar S, Ye J. Drosophila gene expression pattern annotation using sparse features and term-term interactions. Proceedings of the 15th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM, Paris, France. 2009. pp. 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Shattuck D, Damasio H, Leahy R. Geodesic curvature flow on surfaces for automatic sulcal delineation. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI’12; Barcelona, Spain. 2012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Leahy RM, Thompson PM, Shattuck DW. Cortical Surface Parameterization by P-Harmonic Energy Minimization. Biomedical Imaging: From Nano to Macro 2004. ISBI 2004. IEEE International Symposium on; Arlington, VA, USA. 2004. pp. 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Shattuck DW, Thompson PM, Leahy RM. Surface-Constrained Volumetric Brain Registration Using Harmonic Mappings. IEEE Trans Med Imag. 2007;26(12):1657–1669. doi: 10.1109/tmi.2007.901432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SH, Cabeen RP, Joshi AA, Sun B, Dinov I, Narr KL, Toga AW, Woods RP. Diffeomorphic sulcal shape analysis on the cortex. IEEE Trans Med Imaging. 2012b;31(6):1195–1212. doi: 10.1109/TMI.2012.2186975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Stern J, Rehm K, Schaper K, Hurdal MK, Rottenberg D. Cortical Surface Flattening using Least Squares Conformal Mapping with Minimal Metric Distortion. Biomedical Imaging: From Nano to Macro, 2004. ISBI 2004. IEEE International Symposium on; Arlington, VA, USA. 2004. pp. 77–80. [Google Scholar]
- Ju L, Hurdal MK, Stern J, Rehm K, Schaper K, Rottenberg D. Quantitative Evaluation of Three Surface Flattening Methods. NeuroImage. 2005;28(4):869–880. doi: 10.1016/j.neuroimage.2005.06.055. [DOI] [PubMed] [Google Scholar]
- Khosla A, Cao Y, Lin CC-Y, Chiu H-K, Hu J, Lee H. An integrated machine learning approach to stroke prediction. Proceedings of the 16th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM, Washington, DC, USA. 2010. pp. 183–192. [Google Scholar]
- Kim S, Xing EP. Tree-guided group lasso for multi-task regression with structured sparsity. 27th International Conference on Machine Learning; Haifa, Israel. 2010. [Google Scholar]
- Kim WH, Pachauri D, Hatt C, Chung MK, Johnson SC, Singh V. Wavelet based multi-scale shape features on arbitrary surfaces for cortical thickness discrimination. Advances in Neural Information Processing Systems (NIPS) 2012 [PMC free article] [PubMed] [Google Scholar]
- Kindlmann G, Estepar RS, Niethammer M, Haker S, Westin CF. Geodesic-loxodromes for diffusion tensor interpolation and difference measurement. Med Image Comput Comput Assist Interv. 2007;10(Pt 1):1–9. doi: 10.1007/978-3-540-75757-3_1. [DOI] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol Aging. 2010;31(8):1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Stein JL, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM. Boosting power to detect genetic associations in imaging using multi-locus, genome-wide scans and ridge regression. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on (ISBI); Chicago, IL. 2011. pp. 1855–1859. [Google Scholar]
- Kuncheva LI, Rodríguez JJ. Classifier ensembles for fMRI data analysis: an experiment. Magnetic resonance imaging. 2010;28(4):583–593. doi: 10.1016/j.mri.2009.12.021. [DOI] [PubMed] [Google Scholar]
- López M, Ramírez J, Górriz JM, Álvarez I, Salas-Gonzalez D, Segovia F, Chaves R, Padilla P, Gómez-Río M, Initiative AsDN. Principal component analysis-based techniques and supervised classification schemes for the early detection of the Alzheimer’s Disease. Neurocomputing. 2011;74(8):1260–1271. [Google Scholar]
- Lao Z, Shen D, Xue Z, Karacali B, Resnick SM, Davatzikos C. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. Neuroimage. 2004;21(1):46–57. doi: 10.1016/j.neuroimage.2003.09.027. [DOI] [PubMed] [Google Scholar]
- Le Floch E, Lalanne C, Pinel P, Moreno A, Trinchera L, Tenenhaus A, Thirion B, Poline JB, Frouin V, Duchesnay E. Bridging the gap between imaging and genetics: a multivariate statistical investigation. 17th Annual Meeting of the Organization on Human Brain Mapping; Québec City, Canada. 2011. [Google Scholar]
- Leporé N, Brun C, Chou Y-Y, Chiang M-C, Dutton RA, Hayashi KM, Luders E, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors. IEEE Trans Med Imag. 2008;27(1):129–141. doi: 10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li H. Variable selection and regression analysis for graph-structured covariates with an application to genomics. The Annals of Applied Statistics. 2010;4(3):1498–1516. doi: 10.1214/10-AOAS332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chakraborty S, Li F, Liu Y. Bayesian Regularization via the Graph Laplacian Prior. 2010a. [Google Scholar]
- Liu J, Ji S, Ye J. SLEP: Sparse Learning with Efficient Projections. Arizona State University; 2009. http://www.public.asu.edu/~jye02/Software/SLEP. [Google Scholar]
- Liu J, Ye J. Advances in Neural Information Processing Systems. 2010a. Moreau-Yosida Regularization for Grouped Tree Structure Learning. [Google Scholar]
- Liu J, Ye J. Arxiv. 2010b. Fast overlapping group lasso. preprint arXiv:1009.0306. [Google Scholar]
- Liu J, Yuan L, Ye J. An efficient algorithm for a class of fused lasso problems. Proceedings of the 16th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM, Washington, DC, USA. 2010b. pp. 323–332. [Google Scholar]
- MacDonald D. PhD thesis. McGill University; 1998. A method for identifying geometrically simple surfaces from three dimensional images. [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N, Bühlmann P. Stability selection. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010;72(4):417–473. [Google Scholar]
- Memoli F, Sapiro G, Thompson P. Implicit brain imaging. NeuroImage. 2004;23:S179–S188. doi: 10.1016/j.neuroimage.2004.07.072. [DOI] [PubMed] [Google Scholar]
- Michel V, Gramfort A, Varoquaux G, Eger E, Thirion B. Total variation regularization for fMRI-based prediction of behavior. IEEE Trans Med Imaging. 2011;30(7):1328–1340. doi: 10.1109/TMI.2011.2113378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moakher M. A Differential Geometric Approach to the Geometric Mean of Symmetric Positive-Definite Matrices. SIAM J Matrix Anal Appl. 2005;26(3):735–747. [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009;45(1, Supplement 1):S3– S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Toga AW, Thompson PM. Comparison of AdaBoost and Support Vector Machines for Detecting Alzheimer’s Disease Through Automated Hippocampal Segmentation. Medical Imaging, IEEE Transactions on. 2010;29(1):30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov Y. Gradient methods for minimizing composite objective function. ReCALL. 2007;76(2007076) [Google Scholar]
- Ono M, Kubik S, Abernathey CD. Atlas of the cerebral Sulci. Stuttgart, Germany: Thireme; 1990. [Google Scholar]
- Pantazis D, Joshi A, Jiang J, Shattuck DW, Bernstein LE, Damasio H, Leahy RM. Comparison of landmark-based and automatic methods for cortical surface registration. NeuroImage. 2010;49(3):2479–2493. doi: 10.1016/j.neuroimage.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennec X, Fillard P, Ayache N. A Riemannian Framework for Tensor Computing. Int J Comput Vision. 2006;66(1):41–66. [Google Scholar]
- Qiu A, Miller MI. Cortical hemisphere registration via large deformation diffeomorphic metric curve mapping. Med Image Comp Comput-Assist Intervention, Proceedings. 2007:186–193. doi: 10.1007/978-3-540-75757-3_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Eckert MA, Rose FE, Karchemskiy A, Kesler S, Chang M, Reynolds MF, Kwon H, Galaburda A. An experiment of nature: brain anatomy parallels cognition and behavior in Williams syndrome. J Neurosci. 2004;24(21):5009–5015. doi: 10.1523/JNEUROSCI.5272-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryali S, Chen T, Supekar K, Menon V. Estimation of functional connectivity in fMRI data using stability selection-based sparse partial correlation with elastic net penalty. Neuroimage. 2012;59(4):3852–3861. doi: 10.1016/j.neuroimage.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Van Leemput K. The Relevance Voxel Machine (RVoxM): a Bayesian method for image-based prediction. Med Image Comput Comput Assist Interv. 2011;14(Pt 3):99–106. doi: 10.1007/978-3-642-23626-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EL, Shaw A, Wolfson E. A Numerical Solution to the Generalized Mapmaker’s Problem: Flattening Nonconvex Polyhedral Surfaces. IEEE Trans Patt Anal Mach Intell. 1989;11(9):1005–1008. [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic Resonance Image Tissue Classification using a Partial Volume Model. NeuroImage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shen L, Qi Y, Kim S, Nho K, Wan J, Risacher SL, Saykin AJ. Sparse bayesian learning for identifying imaging biomarkers in AD prediction. Med Image Comput Comput Assist Interv. 2010;13(Pt 3):611–618. doi: 10.1007/978-3-642-15711-0_76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Morra JH, Thompson PM, Toga AW. Inverse-Consistent Surface Mapping with Laplace-Beltrami Eigen-Features. Information Processing in Medical Imaging. 2009:467–478. doi: 10.1007/978-3-642-02498-6_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Rex D, Kornsand D, Tessner KD, Jernigan TL, Toga AW. Mapping sulcal pattern asymmetry and local cortical surface gray matter distribution in vivo: maturation in perisylvian cortices. Cereb Cortex. 2002;12(1):17–26. doi: 10.1093/cercor/12.1.17. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6(3):309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Stearns S. On selecting features for pattern classifiers. 3-d International Conference on Pattern Recognition; Coronado, CA. 1976. pp. 71–75. [Google Scholar]
- Stonnington CM, Chu C, Kloppel S, Jack CR, Jr, Ashburner J, Frackowiak RS. Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage. 2010;51(4):1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Medical Image Analysis. 2004;8(3):197– 203. doi: 10.1016/j.media.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci U S A. 2005;102(13):4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Pearlson G, Caprihan A, Adali T, Kiehl KA, Liu J, Yamamoto J, Calhoun VD. Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage. 2011;57(3):839–855. doi: 10.1016/j.neuroimage.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, van Erp TGM, Thompson PM, Bearden CE, Daley M, Kushan L, Hardt ME, Nuechterlein KH, Toga AW, Cannon TD. Elucidating a Magnetic Resonance Imaging-Based Neuroanatomic Biomarker for Psychosis: Classification Analysis Using Probabilistic Brain Atlas and Machine Learning Algorithms. Biological Psychiatry. 2009a;66(11):1055–1060. doi: 10.1016/j.biopsych.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Patel R, Liu J, Chen K, Wu T, Li J, Reiman E, Ye J. Mining brain region connectivity for alzheimer’s disease study via sparse inverse covariance estimation. Proceedings of the 15th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM, Paris, France. 2009b. pp. 1335–1344. [Google Scholar]
- Thirion JP, Prima S, Subsol G, Roberts N. Statistical analysis of normal and abnormal dissymmetry in volumetric medical images. Med Image Anal. 2000;4(2):111–121. doi: 10.1016/s1361-8415(00)00012-8. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Toga AW. A surface-based technique for warping 3-dimensional images of the brain. IEEE Trans Med Imag. 1996;15(4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth Patterns in the Developing Human Brain Detected Using Continuum-Mechanical Tensor Mapping. Nature. 2000a;404(6774):190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/Computational Challenges in Creating Population-Based Brain Atlases. Human Brain Mapping. 2000b;9(2):81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Zubicaray GD, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of Gray Matter Loss in Alzheimer’s Disease. J Neuroscience. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer’s disease. NeuroImage. 2004a;22(4):1754– 1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang Y, van Erp TGM, Cannon TD, Toga AW. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. NeuroImage. 2004b;23(Supplement 1):S2– S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Eckert MA, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Reiss AL. Abnormal Cortical Complexity and Thickness Profiles Mapped in Williams syndrome. J Neuroscience. 2005;25(16):4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang MC, Leow AD, Sowell ER, De Zubicaray G, Becker JT, Lopez OL, Aizenstein HJ, Toga AW. Tracking Alzheimer’s disease. Ann N Y Acad Sci. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996;58(1):267–288. [Google Scholar]
- Tikhonov AN, Arsenin VY. Solutions of Ill Posed Problems. Vh Winston; 1977. [Google Scholar]
- Timsari B, Leahy RM. Optimization method for creating semi-isometric flat maps of the cerebral cortex. Medical Imaging 2000: Image Processing. 2000;3979(1):698–708. [Google Scholar]
- Tosun D, Prince JL. A Geometry-Driven Optical Flow Warping for Spatial Normalization of Cortical Surfaces. Medical Imaging, IEEE Transactions on. 2008;27(12):1739–1753. doi: 10.1109/TMI.2008.925080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant M, Glaunes J. Surface matching via currents. Inf Process Med Imaging. 2005;19:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- Vaillant M, Qiu A, Glaunes J, Miller MI. Diffeomorphic metric surface mapping in subregion of the superior temporal gyrus. Neuroimage. 2007;34(3):1149–1159. doi: 10.1016/j.neuroimage.2006.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An Integrated Software Suite for Surface-based Analyses of Cerebral Cortex. J Am Med Inform Assoc. 2001;8(5):443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Whitwell JL, Kantarci K, Josephs KA, Parisi JE, Shiung MS, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Jack CR., Jr Antemortem MRI based STructural Abnormality iNDex (STAND)-scores correlate with postmortem Braak neurofibrillary tangle stage. NeuroImage. 2008;42(2):559–567. doi: 10.1016/j.neuroimage.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. Neuroimage. 2010;53(3):1147–1159. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vounou M, Janousova E, Wolz R, Stein JL, Thompson PM, Rueckert D, Montana G. Sparse reduced-rank regression detects genetic associations with voxel-wise longitudinal phenotypes in Alzheimer’s disease. Neuroimage. 2012;60(1):700–716. doi: 10.1016/j.neuroimage.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nie F, Huang H, Risacher SL, Saykin AJ, Shen L. A new sparse multi-task regression and feature selection method to identify brain imaging predictors for memory performance. Computer Vision, IEEE 13th International Conference on; ICCV. 2011a. pp. 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nie F, Huang H, Kim S, Nho K, Risacher SL, Saykin AJ, Shen L. Identifying quantitative trait loci via group-sparse multitask regression and feature selection: an imaging genetics study of the ADNI cohort. Bioinformatics. 2012a;28(2):229–237. doi: 10.1093/bioinformatics/btr649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. NeuroImage. 2003;20(2):667– 682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chiang M-C, Thompson PM. Automated Surface Matching using Mutual Information Applied to Riemann Surface Structures. Med. Image Comp. Comput.-Assist. Intervention, Proceedings; 2005. pp. 666–674. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu X, Chan TF, Thompson PM, Yau S-T. Brain Surface Conformal Parameterization with Algebraic Functions. Med Image Comp Comput-Assist Intervention, Proceedings. 2006;Part II:946–954. doi: 10.1007/11866763_116. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lui LM, Gu X, Hayashi KM, Chan TF, Toga AW, Thompson PM, Yau S-T. Brain Surface Conformal Parameterization using Riemann Surface Structure. IEEE Trans Med Imag. 2007;26(6):853–865. doi: 10.1109/TMI.2007.895464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gu X, Chan TF, Thompson PM, Yau S-T. Conformal Slit Mapping and Its Applications to Brain Surface Parameterization. Med Image Comp Comput-Assist Intervention, Proceedings. 2008a;Part I:585–593. doi: 10.1007/978-3-540-85988-8_70. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yin X, Zhang J, Gu X, Chan TF, Thompson PM, Yau S-T. Brain Mapping with the Ricci Flow Conformal Parameterization and Multivariate Statistics on Deformation Tensors. 2nd MICCAI Workshop on Mathematical Foundations of Computational Anatomy; 2008b. pp. 36–47. [Google Scholar]
- Wang Y, Chan TF, Toga AW, Thompson PM. Multivariate Tensor-based Brain Anatomical Surface Morphometry via Holomorphic One-Forms. Med Image Comp Comput-Assist Intervention, Proceedings. 2009a;12(Pt 1):337–344. doi: 10.1007/978-3-642-04268-3_42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gu X, Chan TF, Thompson PM. Shape Analysis with Conformal Invariants for Multiply Connected Domains and its Application to Analyzing Brain Morphology. IEEE Conf Comp Vis Patt Recog CVPR ‘09. 2009b:202–209. [Google Scholar]
- Wang Y, Fan Y, Bhatt P, Davatzikos C. High-dimensional pattern regression using machine learning: from medical images to continuous clinical variables. Neuroimage. 2010a;50(4):1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Senstad R, Toga AW, Thompson PM. MRI-based Biomarker Detection using Conformal Slit Maps and Machine Learning. The 16th Annual Meeting of the Organization for Human Brain Mapping; Barcelona, Spain. 2010b. [Google Scholar]
- Wang Y, Zhang J, Gutman B, Chan TF, Becker JT, Aizenstein HJ, Lopez OL, Tamburo RJ, Toga AW, Thompson PM. Multivariate tensor-based morphometry on surfaces: Application to mapping ventricular abnormalities in HIV/AIDS. NeuroImage. 2010c;49(3):2141–2157. doi: 10.1016/j.neuroimage.2009.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Song Y, Rajagopalan P, An T, Liu K, Chou YY, Gutman B, Toga AW, Thompson PM. Surface-based TBM boosts power to detect disease effects on the brain: An N=804 ADNI study. Neuroimage. 2011b;56(4):1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi J, Yin X, Gu X, Chan TF, Yau S-T, Toga AW, Thompson PM. Brain Surface Conformal Parameterization with the Ricci Flow. IEEE Trans Med Imag. 2012b;31(2):251–264. doi: 10.1109/TMI.2011.2168233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. NeuroImage. 2010;53(3):1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP. Characterizing volume and surface deformations in an atlas framework: theory, applications, and implementation. NeuroImage. 2003;18(3):769–788. doi: 10.1016/s1053-8119(03)00019-3. [DOI] [PubMed] [Google Scholar]
- Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y. Robust Face Recognition via Sparse Representation. IEEE Trans Pattern Anal and Machine Intell. 2009;31(2):210–227. doi: 10.1109/TPAMI.2008.79. [DOI] [PubMed] [Google Scholar]
- Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Xi Y, Hasson U, Ramadge P. Boosting with Spatial Regularization. In: Bengio Y, Schuurmans D, Lafferty J, Williams CKI, Culotta A, editors. Advances in Neural Information Processing Systems. Vol. 22. 2009. pp. 2107–2115. [Google Scholar]
- Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. A Hybrid Machine Learning Method for Fusing fMRI and Genetic Data: Combining both Improves Classification of Schizophrenia. Front Hum Neurosci. 2010;4:192. doi: 10.3389/fnhum.2010.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Shapiro LG, Cunningham ML, Speltz M, Lee S-l. Classification and Feature Selection for Craniosynostosis. ACM Conference on Bioinformatics, Computational Biology & Biomedicine; Chicago, IL. 2011. [Google Scholar]
- Ye J, Farnum M, Yang E, Verbeeck R, Lobanov V, Raghavan N, Novak G, Dibernardo A, Narayan VA. Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. BMC Neurol. 2012;12(1):46. doi: 10.1186/1471-2377-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Sabuncu M, Vercauteren T, Ayache N, Fischl B, Golland P. Spherical demons: fast surface registration. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):745–753. doi: 10.1007/978-3-540-85988-8_89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Sabuncu MR, Vercauteren T, Ayache N, Fischl B, Golland P. Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE Trans Med Imaging. 2010;29(3):650–668. doi: 10.1109/TMI.2009.2030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. Neuroimage. 2012;61(3):622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006;68(1):49–67. [Google Scholar]
- Yushkevich P, Joshi S, Pizer SM, Csernansky JG, Wang LE. Feature selection for shape-based classification of biological objects. Inf Process Med Imaging. 2003;18:114–125. doi: 10.1007/978-3-540-45087-0_10. [DOI] [PubMed] [Google Scholar]
- Zhang T. Analysis of Multi-stage Convex Relaxation for Sparse Regularization. J Mach Learn Res. 2010;11:1081–1107. [Google Scholar]
- Zhao P, Rocha G, Yu B. The composite absolute penalties family for grouped and hierarchical variable selection. Annals of Statistics. 2009;37(6A):3468–3497. [Google Scholar]
- Zhong J, Qiu A. Multi-manifold diffeomorphic metric mapping for aligning cortical hemispheric surfaces. NeuroImage. 2010;49(1):355–365. doi: 10.1016/j.neuroimage.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67(2):301–320. [Google Scholar]
- Zou H. The Adaptive Lasso and Its Oracle Properties. Journal of the American Statistical Association. 2006;101(476):1418–1429. [Google Scholar]