Abstract
It is known that cells under stress accumulate various dinucleoside polyphosphates, compounds suggested to function as alarmones. In plants, the phenylpropanoid pathways yield metabolites protecting these organisms against various types of stress. Observations reported in this communication link these two phenomena and provide an example of a metabolic “addressee” for an “alarm” signaled by diadenosine triphosphate (Ap3A) or diadenosine tetraphosphate (Ap4A). In response to added Ap3A or Ap4A, seedlings of Arabidopsis thaliana incubated in full nutrition medium increased both the expression of the genes for and the specific activity of phenylalanine ammonia-lyase and 4-coumarate:coenzyme A ligase, enzymes that control the beginning of the phenylpropanoid pathway. Neither adenine mononucleotides (AMP, ADP or ATP) nor adenosine evoked such effects. Reactions catalyzed in vitro by these enzymes were not affected by Ap3A or Ap4A.
Keywords: Diadenosine tetraphosphate, Diadenosine triphosphate, Phenylalanine ammonia-lyase, 4-Coumarate:CoA ligase, Phenylpropanoid pathways, Alarmones
Abbreviations: PAL, phenylalanine ammonia-lyase; 4CL, 4-coumarate:coenzyme A ligase; CHS, chalcone synthase; Ap3A, diadenosine 5’,5″′-P1,P3-triphosphate; Ap4A, diadenosine 5′,5″′-P1,P4-tetraphosphate; HPLC, high performance liquid chromatography
1. Introduction
Dinucleoside 5′,5″′-P1,Pn-polyphosphates, NpnN’s (where N and N′ are 5′-O-nucleosides and n is the number of phosphate residues in the polyphosphate chain that links the two 5′-esterified nucleosides), are naturally occurring compounds. They can be synthesized by some ligases [1–6], firefly luciferase [7] and certain transferases [8–10], and have been identified in bacteria [11,12], yeast [13,14] and animals, including sea urchin [15], Artemia salina [16], Drosophila [17] and mammals [18–20]. The presence of NpnN’s has been demonstrated in yellow lupin seedlings (Guranowski, unpublished observation) but no detailed report of the occurrence of these compounds in plants has appeared yet. Three plant ligases, phenylalanyl- and seryl-tRNA synthetases [4] and 4-coumarate:CoA ligase [6], have been shown to catalyze the synthesis of diadenosine 5′,5″′-P1,P4-tetraphosphate (Ap4A) and some other adenylyl derivatives. Since it has been shown that cells subjected to stresses such as elevated temperature, ethanol or cadmium, accumulate various NpnN’s [11–14], these compounds have been termed alarmones. However, no clear metabolic or molecular target (“addressee”) of the postulated “alarm” signalled by the NpnN’s has been experimentally demonstrated. In higher plants, heavy metals including cadmium (II) stimulate the production of many compounds that protect plant tissues against these harmful agents. Among such compounds are products of the phenylpropanoid pathway, including flavonoids and lignins [21–24]. We wondered, therefore, whether exogenously applied Ap3A and Ap4A, the most predominant NpnN’s that probably also accumulate in plant cells subjected to stress-inducing agents like Cd(II), could affect the activities and/or synthesis of any enzymes of the phenylpropanoid pathway. This communication reports that 7-day-old seedlings of Arabidopsis thaliana incubated in medium containing micromolar concentration of Ap3A or Ap4A increased the specific activities of phenylalanine ammonia-lyase (PAL) and 4-coumarate:CoA ligase (4CL) as well as the expression of the genes encoding these enzymes. To the best of our knowledge, this is the first evidence in a plant system that exogenously applied NpnN’s can signal stress conditions by triggering a cascade of reactions to yield various protective compounds.
2. Materials and methods
2.1. Plant growth conditions
Wild-type A. thaliana, ecotype Col-0 (Lehle Seeds, USA) were grown in sterile full nutrition medium prepared as described by Scheible and co-workers [25]. The Arabidopsis seedlings (100–120) were kept in 250-ml glass Erlenmeyer flasks containing 30 ml of the above medium in orbital shakers with constant (24 h), uniform fluorescent light (150 μmol m−2 s−1) at 22 °C. During the first 3 days the shaker’s speed was low (30 rpm) and then it was set at 80 rpm. After 5 days, the old medium was removed and replaced with a fresh portion of the same medium. After a further 2 days each flask was supplemented with a 30 μl aliquot of the compound under study at an appropriate concentration, or water (control), and the growth continued. The plants were harvested after 5, 10, 30, 60, 120 or 180 min, depending on the experiment. A group of plants from each flask was quickly blotted on tissue paper, washed twice with an excess of distilled water, blotted on tissue paper again, frozen in liquid nitrogen, and kept at −80 °C for analysis.
2.2. Determination of Ap3A and Ap4A in the growth medium
To monitor uptake of the dinucleotides by A. thaliana seedlings, samples of the growth medium (2 ml) were collected at the same time as the plants and analyzed by HPLC as described earlier [26].
2.3. mRNA level determination
Total RNA was extracted from A. thaliana seedlings using an RNeasy Plant Kit (Qiagen). DNA was removed with RNase-free DNase (Qiagen). RNA purity was confirmed by PCR using actin-specific primers. RNA concentration was determined with a Qubit fluorometer (Invitrogen) and 4 μg of total RNA was used for cDNA synthesis. RNA and oligo(dT)19 (50 μM) primers were mixed in a total volume of 42 μl and incubated for 5 min at 65 °C followed by 1 min on ice. SuperScript III reverse transcriptase (Invitrogen), dNTP mix, 5× first strand buffer, DTT and RNase inhibitor (RNaseOUT Invitrogen) were mixed at 4 °C and dispensed into the tubes with RNA. The reaction was carried out in 60 μl at 50 °C for 60 min. Reverse transcriptase was inactivated by heating at 70 °C for 15 min. A real-time quantitative PCR reaction was performed (Mastercycler® ep realplex, Eppendorf) on the synthesized cDNA (20 ng) using HotStar-IT SYBR Green qPCR Master Mix (USB) and the following primers specific for A. thaliana (PAL1, PAL2, 4CL, common to 4CL1, 4CL2 and 4CL3, and CHS, respectively): PAL1F 5′-CCAAATGATTGTCTGTGAAGTGG-3′, PAL1R 5′-CCGATGTTTGTTATGGATATTGAG-3′, PAL2F 5′-CAATGGATCAAATCGAAGCA-3′, PAL2R 5′-TATTCCGGCGTTCAAAAATC-3′ and 4CL (4CL1, 4CL2, and 4CL3); 4CLF 5′-CATCCCTAACCACCTCCCACTC-3′ i 4CLR 5′-GGAGGAGGATCATTACAACGTC-3′, CHSF 5′-GGCAAAGAAGCGGCAGTGAAGG-3′ and CHSR 5′-GACGGAAGGACGGAGACCAAG-3′. Standard cycling conditions were: 2 min at 50 °C, 10 min at 95 °C and 40 cycles altering between 15 s at 95 °C and 55 °C for 15 s and 1 min at 60 °C, then the melting curve profiles were determined. The comparative CT (cycle threshold) method for relative quantification was used with actin ACTF 5′-ACTTTCATCAGCCGTTTTGA-3′ and ACTR 5′-ACGATTGGTTGAATATCATCAG-3’ as the endogenous control. The amount of target, normalized to an endogenous reference and relative to the calibrator, was determined using the method [27]. The GenBank Accession Nos. for the sequences used in this work are: NM_129260 (PAL1), NM_115186 (PAL2), NM_179462 (4CL1), NM_113019 (4CL2), NM_179513 (4CL3), NM_121396 (CHS), NM_114519 (actin).
2.4. Enzyme extraction and assays
2.4.1. PAL activity
Frozen A. thaliana seedlings (0.5 g) were ground in a mortar in liquid nitrogen and mixed with 5 ml of extraction buffer (150 mM Tris–HCl, pH 8.8, 12 mM 2-mercaptoethanol, 0.1 g ml−1 Dowex® 1X4-200). The homogenate was centrifuged at 23,000g for 30 min at 4 °C. The supernatant, referred to as the enzyme extract, was used for the determination of phenylalanine ammonia-lyase (EC 4.3.1.5) activity according to Alokam and co-workers [28] by measuring the increase in A290 for 10 min at 30 °C due to the accumulation of trans-cinnamic acid (ε290 = 9.5 mM−1 cm−1). The PAL assay mixture (0.2 ml) contained 50 mM Tris–HCl, pH 8.8, 5 mM l-phenylalanine and 10 μl enzyme extract.
2.4.2. 4CL activity
Frozen A. thaliana seedlings (0.5 g) were ground in a mortar with liquid nitrogen and 5 ml of extraction buffer (100 mM Tris–HCl, pH 7.8, 5 mM 2-mercaptoethanol, 5% glycerol). Next, 0.5 g Dowex® 1X4-200 was added and the sample stirred for 15 min at 4 °C. The homogenate was centrifuged at 23,000g for 30 min. The 4:coumarate-CoA ligase (EC 6.2.1.12) was assayed according to Knobloch and Hahlbrock [29]. The reaction mixture (0.2 ml) contained 100 mM Tris–HCl (pH 7.8), 0.1 mM p-coumaric acid, 0.5 mM ATP, 0.3 mM CoA, 5 mM MgCl2 and 10 μl enzyme extract (4-6 μg of protein). The activity of 4CL was determined at 30 °C. Formation of coumaroyl-CoA was measured by monitoring the A333 (ε333 = 21 mM−1 cm−1) [30]. Assays with Ap3A or Ap4A were performed with pure recombinant At4CL2 kindly donated by Dr. Erich Kombrink (Max Planck Institute for Plant Breeding Research, Cologne, Germany).
2.4.3. CHS activity
The extraction and assay of chalcone synthase (EC 2.3.1.74) was performed according to a modification of the method of Fischer and co-workers [31]. Frozen A. thaliana seedlings (0.5 g) were ground in a mortar in liquid nitrogen and mixed with extraction buffer (100 mM KH2PO4/K2HPO4 pH 8.0, 18 mM l-cysteine, 20 mM ascorbic acid, 0.1 g ml−1 Dowex® 1X4-200). The homogenate was centrifuged at 23,000 g for 30 min and the supernatant (enzyme extract) used for the enzyme assay. The reaction mixture (63 μl) contained 50 mM KH2PO4/K2HPO4 pH 8.0, 20 mM l-cysteine, 2% BSA (w/v), 0.2 mM p-coumaroyl-CoA (see below for its synthesis), 0.2 mM [2-14C]malonyl-CoA and 10 μl enzyme extract (4–6 μg of protein). Incubation was carried out at 35 °C for 1 h. After this time, the reaction was stopped by adding 6 μl 20% HCl. Next, 200 μl ethyl acetate was added and the reaction mixed using a vortex and centrifuged for at least 2 min. The ethyl acetate layer was transferred to a new tube and evaporated to dryness in a SpeedVac concentrator. The sample was redissolved in 20 μl methanol and applied to an aluminum-backed silica gel plate containing a fluorescent indicator (Merck, Cat. No. 5554). The chromatogram was developed for 45 min in chloroform:ethanol (3:1 vol/vol), dried, and the naringenin visualized under a short-wave ultraviolet lamp. The spots of naringenin were cut out and radioactivity determined by scintillation counting.
p-Coumaroyl-CoA was prepared according to Sullivan [32] using recombinant At4CL2 protein. The thioester was synthesized in a 2-ml reaction mixture containing 100 mM Tris-HCl pH 7.8, 0.5 mM CoA, 5 mM ATP, 5 mM MgCl2, 1 mM p-coumaric acid and 4 μg recombinant 4CL2. The reaction mixture was incubated at 37 °C and monitored by measuring the A333 up to 2 h. Next, the reaction mixture was applied to a 1-ml ENVI-18 solid-phase extraction column (Supelco) preequilibrated with 3 ml methanol and 3 ml 0.1% acetic acid in water, pH 2.75. The column was washed with 6 ml 0.1% acetic acid in water, pH 2.75, and the coumaroyl-CoA eluted with 1 ml methanol. After methanol evaporation, the thioester was dissolved in 0.5 ml 25 mM MOPS buffer, pH 7.5. The concentration of p-coumaroyl-CoA was determined spectrophotometrically (see Section 2.4.2).
2.5. Protein concentration
Total protein concentration was estimated according to Bradford [33], using bovine serum albumin as a standard.
2.6. Statistical analysis
The experiments were carried out in triplicate for the enzyme assays and in duplicate for the mRNA level determination. The results are the mean ± SD.
3. Results
In initial experiments designed to determine whether exogenously applied Ap3A or Ap4A could affect the synthesis and activity of PAL and 4CL in A. thaliana seedlings, a concentration of 5 μM dinucleotide was used in the growth medium. Under these conditions we observed a dramatic time-dependent increase in the expression of the PAL2 gene with either dinucleotide (Fig. 1A). Ap4A triggered an increase in PAL2 expression within the first 5 min of the incubation, reaching a maximum 75-fold increase within 10 min and remaining at this level for at least 3 h. Although we analyzed fewer time points with Ap3A, this nucleotide appeared to cause a very similar effect. Interestingly, PAL1 expression remained practically unaffected. Fig. 1B shows that the specific activity of PAL in seedling extracts also increased in response to Ap3A (up to 8- to 9-fold in 3 h) and Ap4A, though to a lesser extent and with different kinetics (up to 3-fold by 10 min followed by a decline). The dependence of PAL catalytic activity in the seedling extracts on the concentration of Ap3A or Ap4A in the growth medium is shown in Fig. 1C. With Ap3A, a kind of saturation curve can be seen that reaches a plateau at 0.2 μM, decreasing slightly thereafter up to 25 μM. In the case of Ap4A, the highest increase in PAL activity was observed at 40 nM. The magnitude of the response to 8 nM Ap3A is quite striking.
Fig. 1.
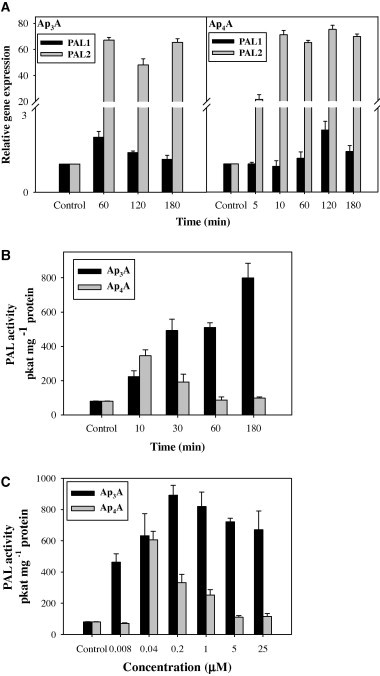
Phenylalanine ammonia-lyase gene PAL1 and PAL2 expression (A) and PAL activity (B and C) in Arabidopsis thaliana seedlings treated with Ap3A or Ap4A. (A) Treatment with 5 μM Ap3A or Ap4A for 0–180 min; (B) treatment with 5 μM Ap3A or Ap4A for 0–180 min; (C) Treatment with 0–25 μM Ap3A or Ap4A for 180 min. Values are means of three independent experiments ±SD.
Analogous measurements were performed for the Arabidopsis 4CL genes (Fig. 2A) and 4CL catalytic activity (Fig. 2B and C). Ap3A- and Ap4A-stimulated expression of the 4CL genes was also observed, although it was much less dramatic (less than 3-fold) than that of PAL2. In the case of 4CL activity, the effects evoked by Ap3A and Ap4A were similar. The plateau of 4CL activity in response to 5 μM Ap3A or Ap4A was reached in 30 min, with that particular concentration of the dinucleotides appearing to exert the strongest effect (Fig. 2C). Finally, we found that the gene and the catalytic activity of chalcone synthase, which catalyzes a more downstream reaction in the phenylpropanoid pathway than does PAL or 4CL, were both poorly stimulated by either AP3A or AP4A (Fig. 3A and B).
Fig. 2.
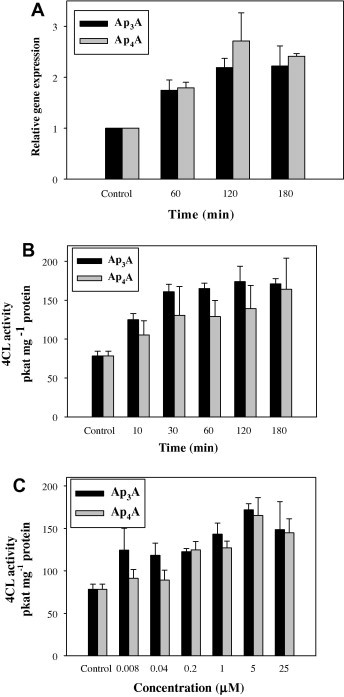
4-Coumarate:CoA ligase gene 4CL expression (A) and 4CL activity (B and C) in Arabidopsis thaliana seedlings treated with Ap3A or Ap4A. (A) Treatment with 5 μM Ap3A or Ap4A for 0–180 min; (B) treatment with 5 μM Ap3A or Ap4A for 0–180 min; (C) treatment with 0–25 μM Ap3A or Ap4A for 180 min. Values are means of three independent experiments ±SD.
Fig. 3.
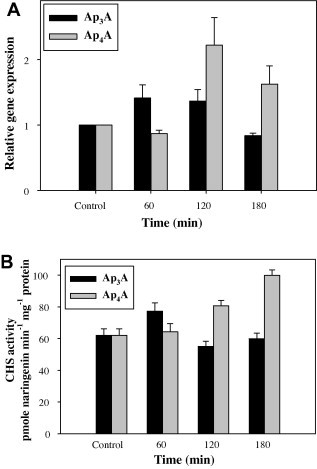
Expression of chalcone synthase gene CHS (A) and CHS activity (B) in Arabidopsis thaliana seedlings treated with Ap3A or Ap4A. (A) Treatment with 5 μM Ap3A or Ap4A for 0–180 min; (B) treatment with 5 μM Ap3A or Ap4A for 0–180 min. Values are means of three independent experiments ±SD.
We also found that neither Ap3A nor Ap4A affected PAL or 4CL activity directly when added at concentrations up to 50 μM to the in vitro assay mixtures (not shown). Since all organisms [34], including plants [35] possess a number of specific and nonspecific enzymes that can degrade NpnN’s yielding nucleoside mononucleotides, we checked whether these potential degradation products (ATP, ADP, AMP or adenosine), used at the same micromolar concentrations as those of Ap3A or Ap4A, could exert the above effects on the genes or activities of PAL and 4CL. When individually tested at a concentration of 5 μM, none of these four compounds could stimulate the expression or activity of either enzyme in the seedling extracts. Using HPLC, we also monitored changes in the concentration of Ap3A and Ap4A in the growth medium during the experiments. In each case, the starting concentration of 5 μM fell to about 3 μM after 3 h but no measurable amounts of adenine mononucleotides were detected in the medium. Thus, the apparent “consumption” of these dinucleotides by the seedlings was not accompanied by the appearance of their potential degradation products in the growth medium.
4. Discussion
For more than three decades, researchers have carried out many different kinds of experiments in different systems to try to answer the question about the biological role of NpnN’s. For example, Ap4A was found to trigger the initiation of DNA replication in vitro [37] and to be a ligand of a 57-kDa protein associated with DNA polymerase α [38]. It also stimulated DNA synthesis when microinjected into Xenopus laevis oocytes [39] and induced apoptosis in cultured human cells [40]. Extracellularly, different ApnNs can control blood pressure [41,42] and act as neurotransmitters [43]. Recently, Ap4A has been reported to be a signaling molecule in immunologically activated mast cells [44]. For more examples and a comprehensive discussion on these issues see the review by McLennan [36]. Until now however, no potential function for NpnN’s has been demonstrated in plants. Our findings described above show that at least Ap3A and Ap4A can act as alarmones in plants. They evoked the strongest response in stimulating the expression and activity of PAL, the enzyme that catalyzes the first reaction of the phenylpropanoid pathways. The response of 4CL was much weaker and that of chalcone synthase was insignificant. It is generally believed that the products of the phenylpropanoid pathways protect plants against various stresses caused by wounding, pathogen infection, ultraviolet irradiation and heavy metals, including cadmium [21,45,46]. A large number of studies have shown that PAL expression is responsive to these environmental stimuli [45,47,48]. PAL activity is a key factor in the increased accumulation of flavonoids and other phenolic compounds under UV-B radiation and water deficit [49,50]. We have checked the expression of PAL1 and PAL2 because these genes proved to be important for lignin synthesis and also have functional specialization in abiotic environmentally-triggered flavonoid synthesis [51]. Expression of various 4CLs, including 4CL1, 4CL2 and 4CL3, and the 4CL activity were also shown to respond to different stresses, in particular to pathogen-related elicitor treatment [52,53] and UV-B irradiation [50,54,55]. Our findings suggest that plant tissues possess a specific receptor that recognizes diadenosine tri- and/or tetraphosphates but not adenine mononucleotides and apparently triggers a cascade of events to yield these protective metabolites. Thus our work opens up new avenues for studies on the role of Ap3A, Ap4A and of other NpnN′s in plants. In the near future, efforts should be undertaken to at least answer such questions as: (i) do the non-adenylylated NpnN′s evoke the same effects as Ap3A or Ap4A; (ii) is this phenomenon common to other plant systems; (iii) can a plant cell receptor be identified with specificity for these dinucleotides; (iv) do the exogenously applied diadenosine polyphosphates affect accumulation of particular phenylpropanoic compound(s) in the plant tissues; and (v) how do other genes and enzymes of the phenylpropanoid pathways respond to those uncommon (di)nucleotides?
Based on existing knowledge of the reactions caused in cells by cadmium [12,13,21–23] and on the observations communicated in this paper, we postulate that in plant cells Cd (II) causes accumulation of Ap3A and/or Ap4A and, by analogy with the activation of the MITF transcription factor in mast cells by Ap4A [44], these compounds interact with transcription factors that control mainly the PAL2 gene and to a lesser extent the 4CL genes. Since the metabolites of the phenylpropanoid pathways protect plants against the harmful effects of different types of stress, Ap3A and Ap4A behave in our biological system as true alarmones, initiating the rescue action. Finally, as 4CL is considered to be an enzyme involved in the response to different harmful factors, one can hypothesize that it plays at least two roles under conditions of stress. First, it is able to synthesize the diadenosine polyphosphate (the putative alarmones) [6] and secondly, as one of the enzymes of the phenylpropanoid pathways, it then contributes to the production of metabolites that minimize the effects of the stress.
Note: Preliminary report of this study was presented as a poster at the 46th Meeting of the Polish Biochemical Society (Cracow, September 5–9, 2011) [56].
Acknowledgements
This study was supported by the Ministry of Science and Higher Education (Grant No. N N303 068634). We thank Professor Alexander McLennan (University of Liverpool, UK) for critical reading of the manuscript and linguistic help in preparation of this paper.
Contributor Information
Małgorzata Pietrowska-Borek, Email: gospi@up.poznan.pl.
Andrzej Guranowski, Email: guranow@up.poznan.pl.
References
- 1.Zamecnik P.C., Stephenson M.L., Janeway C.M., Randerath K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 1966;24:91–97. doi: 10.1016/0006-291x(66)90415-3. [DOI] [PubMed] [Google Scholar]
- 2.Plateau P., Mayaux J.-F., Blanquet S. Zinc (II)-dependent synthesis of diadenosine 5′, 5″′-P1,P4-tetraphosphate by Escherichia coli and yeast phenylalanyl transfer ribonucleic acid synthetases. Biochemistry. 1981;20:4654–4662. doi: 10.1021/bi00519a021. [DOI] [PubMed] [Google Scholar]
- 3.Goerlich O., Foeckler R., Holler E. Mechanism of synthesis of adenosine(5′)tetraphospho(5′)adenosine (AppppA) by aminoacyl-tRNA synthetases. Eur. J. Biochem. 1982;126:135–142. doi: 10.1111/j.1432-1033.1982.tb06757.x. [DOI] [PubMed] [Google Scholar]
- 4.Jakubowski H. Synthesis of diadenosine 5′, 5″′-P1,P4-tetraphosphate and related compounds by plant (Lupinus luteus) seryl-tRNA and phenylalanyl-tRNA synthetases. Acta Biochim. Pol. 1983;30:51–69. [PubMed] [Google Scholar]
- 5.Fontes R., Günther Sillero M.A., Sillero A. Acyl-coenzyme A synthetase from Pseudomonas fragi catalyzes the synthesis of adenosine 5′-polyphosphates and dinucleoside polyphosphates. J. Bacteriol. 1998;180:3152–3158. doi: 10.1128/jb.180.12.3152-3158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietrowska-Borek M., Stuible H.P., Kombrink E., Guranowski A. 4-Coumarate: coenzyme A ligase has the catalytic capacity to synthesize and reuse various (di)adenosine polyphosphates. Plant Physiol. 2003;131:1401–1410. doi: 10.1104/pp.011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guranowski A., Günther Sillero M.A., Sillero A. Firefly luciferase synthesizes P1,P4-bis(5′-adenosyl) tetraphosphate (Ap4A) and other dinucleoside polyphosphates. FEBS Lett. 1990;271:215–218. doi: 10.1016/0014-5793(90)80409-c. [DOI] [PubMed] [Google Scholar]
- 8.Guranowski A., Just G., Holler E., Jakubowski H. Synthesis of diadenosine 5′, 5″′-P1,P4-tetraphosphate (AppppA) from adenosine 5′-phosphosulfate and adenosine 5′-triphosphate catalyzed by yeast AppppA phosphorylase. Biochemistry. 1988;27:2959–2964. doi: 10.1021/bi00408a044. [DOI] [PubMed] [Google Scholar]
- 9.Guranowski A., de Diego A., Sillero A., Günther Sillero M.A. Uridine 5′-polyphosphates (p4U and p5U) and uridine(5′)polyphospho(5′)nucleosides (UpnNs) can be synthesized by UTP:glucose-1-phosphate uridylyltransferase from Saccharomyces cerevisiae. FEBS Lett. 2004;561:83–88. doi: 10.1016/S0014-5793(04)00126-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang D., Shatkin A.J. Synthesis of Gp4N and Gp3N compounds by guanylyltransferase purified from yeast. Nucleic Acids Res. 1984;12:2303–2315. doi: 10.1093/nar/12.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee P.C., Barry R., Ames B.N. Diadenosine 5′, 5″′-P1,P4-tetraphosphate and related adenylylated nucleotides in Salmonella typhimurium. J. Biol. Chem. 1983;258:6827–6834. [PubMed] [Google Scholar]
- 12.Coste H., Brevet A., Plateau P., Blanquet S. Non-adenylylated bis(5′-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 1987;262:12096–12103. [PubMed] [Google Scholar]
- 13.Pàlfi Z., Surànyi G., Borbély G. Alterations in the accumulation of adenylated nucleotides in heavy-metal-ion-stressed and heat-stressed Synechococcus sp. Strain PCC 6301, cyanobacterium, in light and dark. Biochem. J. 1991;276:487–491. doi: 10.1042/bj2760487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baltzinger M., Ebel J.-P., Remy P.M. Accumulation of dinucleoside polyphosphates in Saccharomyces cerevisiae under stress conditions: High levels are associated with cell death. Biochimie. 1986;68:1231–1236. doi: 10.1016/s0300-9084(86)80069-4. [DOI] [PubMed] [Google Scholar]
- 15.Morioka M., Shimada H. Synthesis of diadenosine 5′, 5″′-P1,P4-tetraphosphate (AP4A) in sea urchin embryos. Cell Differentiation. 1984;14:53–58. doi: 10.1016/0045-6039(84)90008-3. [DOI] [PubMed] [Google Scholar]
- 16.Miller D., McLennan A.G. Changes in intracellular levels of Ap3A and Ap4A in cysts and larvae of Artemia do not correlate with changes in protein synthesis after heat-shock. Nucleic Acids Res. 1986;14:6031–6040. doi: 10.1093/nar/14.15.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brevet A., Plateau P., Best-Belpomme M., Blanquet S. Variation of Ap4A and other dinucleoside polyphosphates in stressed Drosophila cells. J. Biol. Chem. 1985;260:15566–15570. [PubMed] [Google Scholar]
- 18.Flodgaard H., Klenow H. Abundant amounts of diadenosine 5′, 5″′-P1,P4-tetraphosphate are present and releasable, but metabolically inactive, in human platelets. Biochem. J. 1982;208:737–742. doi: 10.1042/bj2080737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lűthje J., Olivie A. The presence of diadenosine 5′, 5″′-P1,P3-triphosphate (Ap3A) in human platelets. Biochem. Biophys. Res. Commun. 1983;115:253–260. doi: 10.1016/0006-291x(83)90997-x. [DOI] [PubMed] [Google Scholar]
- 20.Garrison P.N., Barnes L.D. Determination of dinucleoside polyphosphates. In: McLennan A.G., editor. Ap4A and Other Dinucleoside Polyphosphates. CRC Press; Boca Raton, FL: 1992. pp. 29–61. [Google Scholar]
- 21.Schützendübel A., Schwanz P., Teichman T., Gross K., Langenfeld-Heyser R., Godbold D.L., Polle A. Cadmium-induced changes in antioxidative systems, hydrogen peroxide content, and differentiation in Scots pine roots. Plant Physiol. 2001;127:887–898. doi: 10.1104/pp.010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai L.-P., Xiong Z.-T., Huang Y., Li M.-J. Cadmium-induced changes in pigments, total phenolics, and phenylalanine ammonia-lyase activity in fronds of Azolla imbricata. Environ. Toxicol. 2006;21:505–512. doi: 10.1002/tox.20212. [DOI] [PubMed] [Google Scholar]
- 23.Sivaci A., Sivaci E.R., Sökmen M. Changes in antioxidant activity, total phenolic and abscisic acid constituents in the aquatic plants Myriophyllum spicatum L. and Myriophyllumtriphyllum Orchard exposed to cadmium. Ecotoxicology. 2007;16:423–428. doi: 10.1007/s10646-007-0145-1. [DOI] [PubMed] [Google Scholar]
- 24.Kováčik J., Klejdus B., Hedbavny J., Zoń J. Significance of phenols in cadmium and nickel uptake. J. Plant Physiol. 2011;168:576–584. doi: 10.1016/j.jplph.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Scheible W.-R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rojas N., Schindelasch D., Thimm O., Udvardi M.K., Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guranowski A., Starzyńska E., Pietrowska-Borek M., Rejman D., Blackburn G.M. Novel diadenosine polyphosphate analogs with oxymethylene bridges replacing oxygen in the polyphosphate chain. Potential substrates and/or inhibitors of Ap4A hydrolases. FEBS J. 2009;276:1546–1553. doi: 10.1111/j.1742-4658.2009.06882.x. [DOI] [PubMed] [Google Scholar]
- 27.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by comparative CT method. Nat. Protoc. 2008;3:1101–1107. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Alokam S., Li Y., Li W., Chinnappa C.C., Reid D.M. Photoregulation of phenylalanine ammonia-lyase (PAL) in the accumulation of anthocyanin in alpine and prairie ecotypes of Stellaria longipes under varied R/FR. Physiol. Plant. 2002;116:531–538. [Google Scholar]
- 29.Knobloch K.H., Hahlbrock K. 4-Coumarate:CoA ligase from cell suspension of Petroselinum hortense Hoffm. Partial purification, substrate specificity, and further properties. Arch. Biochem. Biophys. 1977;184:237–248. doi: 10.1016/0003-9861(77)90347-2. [DOI] [PubMed] [Google Scholar]
- 30.Stöckigt J., Zenk M.H. Chemical syntheses and properties of hydroxycinnamoyl coenzyme A derivatives. Z. Naturforsch. 1975;30c:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- 31.Fischer S., Böttcher U., Reuber S., Anhalt S., Weissenböck G. Chalcone synthase in the liverwort Marchantia polymorpha. Phytochemistry. 1995;39:1007–1012. [Google Scholar]
- 32.Sullivan M.L. A novel red clover hydroxycinnamoyl transferase has enzymatic activities consistent with a role in phaselic acid biosynthesis. Plant Physiol. 2009;150:1866–1879. doi: 10.1104/pp.109.136689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:48–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 34.Guranowski A. Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol. Ther. 2000;87:117–139. doi: 10.1016/s0163-7258(00)00046-2. [DOI] [PubMed] [Google Scholar]
- 35.Jakubowski H., Guranowski A. Enzymes hydrolyzing ApppA and/or AppppA in higher plants; purification and some properties of diadenosine triphosphatase, diadenosine tetraphosphatase, and phosphodiesterase from yellow lupin (Lupinus luteus) seeds. J. Biol. Chem. 1983;258:9982–9989. [PubMed] [Google Scholar]
- 36.McLennan A. Dinucleoside polyphosphates — friend or foe? Pharmacol. Ther. 2000;87:73–89. doi: 10.1016/s0163-7258(00)00041-3. [DOI] [PubMed] [Google Scholar]
- 37.Grummt F. Diadenosine 5′, 5″′-P1,P4-tetraphosphate triggers initiation of in vitro DNA replication in baby hamster kidney cells. Proc. Natl. Acad. Sci. USA. 1978;75:371–374. doi: 10.1073/pnas.75.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grummt F., Waltl G., Jantzen H.-M., Hamprecht K., Huebscher U., Kuenzle C.C. Diadenosine 5′, 5″′-P1,P4-tetraphosphate, a ligand of the 57-kilodalton subunit of DNA polymerase α. Proc. Natl. Acad. Sci. USA. 1979;76:6081–6085. doi: 10.1073/pnas.76.12.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zourgui L., Tharaud D., Solari A., Litvak S., Tarrago-Litvak L. Stimulation of DNA synthesis by microinjection of diadenosine 5′, 5″′-P1,P4-tetraphosphate (Ap4A) into Xenopus laevis oocytes. Dev. Biol. 1984;103:409–413. doi: 10.1016/0012-1606(84)90328-2. [DOI] [PubMed] [Google Scholar]
- 40.Vartanian A., Alexandrov I., Prudovski I., McLennan A., Kisselev L. Ap4A induces apoptosis in cultured human cells. FEBS Lett. 1999;456:175–180. doi: 10.1016/s0014-5793(99)00956-4. [DOI] [PubMed] [Google Scholar]
- 41.Schlűter H., Offers E., Bruggemann G., van der Giet M., Tepel M., Nordhoff E., Karas M., Spieker C., Witzel H., Zidek W. Diadenosine phosphates and the physiological control of blood pressure. Nature. 1994;367:186–188. doi: 10.1038/367186a0. [DOI] [PubMed] [Google Scholar]
- 42.Jankowski V., Tőlle M., Vanholder R., Schőnfelder G., van der Giet M., Henning L., Schlűter H., Paul M., Zidek W., Jankowski J. Uridine adenosine tetraphosphate: a novel endothelium-derived vasoconstrictive factor. Nat. Med. 2005;11:223–227. doi: 10.1038/nm1188. [DOI] [PubMed] [Google Scholar]
- 43.Pintor J., Miras-Portugal M.T. Diadenosine polyphosphates (ApxA) as new neurotransmitters. Drug Dev Res. 1993;28:259–262. [Google Scholar]
- 44.Carmi-Levy I., Motzik A., Ofir-Birin Y., Yagil Z., Yang Z., Yang C.M., Kemeny D.M., Han J.M., Kim S., Kay G., Nechushtan H., Suzuki R., Rivera J., Razin E. Importin beta plays an essential role in the regulation of the LysRS-Ap(4)A pathway in immunologically activated mast cells. Mol. Cell. Biol. 2011;2011:2111–2121. doi: 10.1128/MCB.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon R.A., Paiva N.L. Stress-induced phenylopropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrer J.L., Austin M.B., Stewart C., Jr, Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawton M.A., Dixon R.A., Hahlbrock K., Lamb C. Rapid induction of the synthesis of phenylalanine ammonia-lyase and of chalcone synthase in elicitor-treated plant cells. Eur. J. Biochem. 1983;129:593–601. doi: 10.1111/j.1432-1033.1983.tb07090.x. [DOI] [PubMed] [Google Scholar]
- 48.Olsen M.K., Lea U.S., Slimestad R., Verheul M., Lillo C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008;165:1491–1499. doi: 10.1016/j.jplph.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Liu L., McClure J.W. Effects of UV-B on activities of enzymes of secondary phenolic metabolism in barley primary leaves. Physiol. Plant. 1995;93:734–739. [Google Scholar]
- 50.H. Bandurska, M. Pietrowska-Borek, M. Cieślak, Response of barley seedlings to water deficit and enhanced UV-B irradiation acting alone and in combination. Acta Physiol. Plant, 2011. doi:10.1007/s11738-011-0814-9.
- 51.Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y.-H., Yu J.-Q., Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindermayr C., Möllers B., Fliegmann J., Uhlmann A., Lottspeich F., Meimberg H., Ebel J. Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur. J. Biochem. 2002;269:1304–1315. doi: 10.1046/j.1432-1033.2002.02775.x. [DOI] [PubMed] [Google Scholar]
- 53.Gaid M.M., Scharnhop H., Ramadan H., Beuerle T., Beerhues L. 4-Coumarate:CoA ligase family members from elicitor-treated Sorbus aucuparia cell cultures. J. Plant Physiol. 2011;168:944–951. doi: 10.1016/j.jplph.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 54.Douglas C., Hoffmann H., Schulz W., Hahlbrock K. Structure and elicitor or uv-light stimulated expression of two 4-coumarate:CoA ligase genes in parsley. EMBO J. 1987;6:1189–1195. doi: 10.1002/j.1460-2075.1987.tb02353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura M., Yamamoto Y.Y., Seki M., Sakuari T., Sato M., Abe T., Yoshida S., Manabe K., Shinozaki K., Matsui M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. Pietrowska-Borek, M., Nuc, K., Zielezińska, M., Guranowski, A., 2011. Diadenosine polyphosphates (Ap3A and Ap4A), putative alarmones, trigger synthesis of enzymes of the phenylpropanoid pathway in Arabidopsis thaliana. Acta Biochim. Pol. 58 (Suppl. 2, P27.4) 264. [DOI] [PubMed] [Google Scholar]
- 56.Pietrowska-Borek M., Nuc K., Zielezińska M., Guranowski A. Diadenosine polyphosphates (Ap3A and Ap4A), putative alarmones, trigger synthesis of enzymes of the phenylpropanoid pathway in Arabidopsis thaliana. Acta Biochim. Pol. 2011;58(Suppl. 2. P27.4):264. doi: 10.1016/j.fob.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


