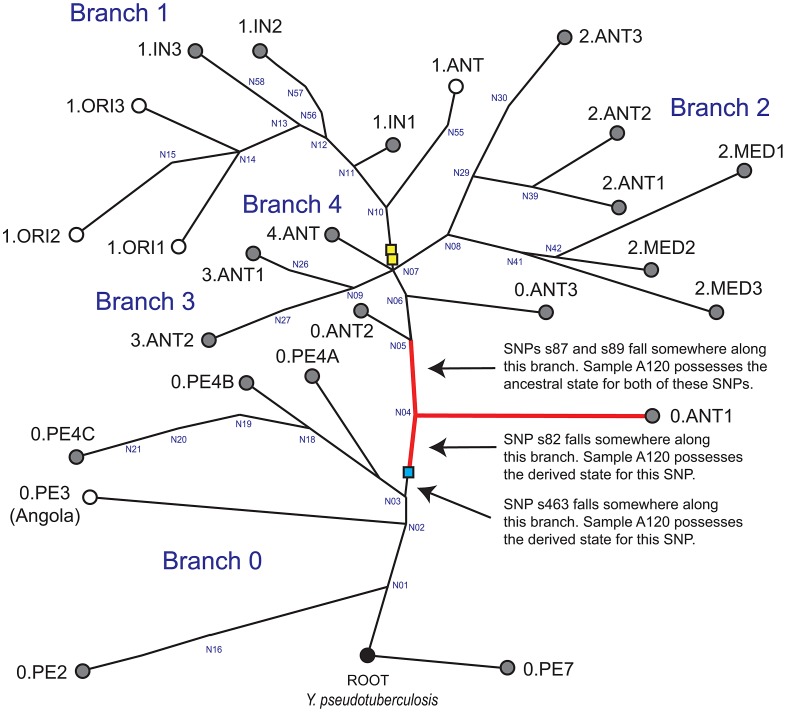
Figure 1. Global phylogeny for Y. pestis.
This global phylogeny for Y. pestis is based upon figures 1A and S3B in Cui et al. [11]. It includes four major branches (0–4) and is rooted with Y. pseudotuberculosis, the ancestor of Y. pestis [28]. The identities of many of the major nodes defined by Cui et al. [11] are presented in blue text. Circles represent specific populations; populations highlighted in gray have, to date, only been found in Asia [10], [11]. Note that the location where strain Angola was isolated, which is the sole representative of population 0.PE3, is unknown. The phylogenetic position of Mongolian strain MNG 2972 is indicated with the blue box (see text). Five previously identified [10], [11] key SNPs were utilized in the current study: s545, which occurs along the branch between nodes N06 and N07 (not shown); s87 and s89; which occur along the branch between N04 and N05; s82, which occurs along the branch between the branching point of strain MNG 2972 and N04; and s463, which occurs along the branch between the branching point of strain MNG 2972 and N03. It is known that these SNPs occur along these specific branches but the exact position and order of these SNPs along each branch is unknown. Sample A120 possesses ancestral states for SNPs s87, s89, and s545; and derived states for SNPs s82 and s463. Thus, the position of sample A120 in this phylogeny is along the branch between the branching point of strain MNG 2972 and N04, along branch N04-N05, along the branch from N04 to 0.ANT1 (red branches), or along one of the sub-branches within 0.ANT.1 (not shown). The phylogenetic positions of strains from the second pandemic [3], [12] are indicated with the yellow boxes according to Cui et al. [11]. The basal node for the 1.ORI group, which caused the third pandemic, is N14 [11].