Abstract
Erwinia amylovora is responsible for fire blight, a necrotic disease of apples and pears. E. amylovora relies on a type III secretion system (T3SS) to induce disease on host plants.
DspA/E belongs to the AvrE family of type III effector. Effectors of the AvrE family are injected via the T3SS in plant cell and are important to promote bacterial growth following infection and to suppress plant defense responses. Their mode of action in the plant cells is unknown. Here we study the physiological effects induced by dspA/E expression in the yeast Saccharomyces cerevisiae. Expression of dspA/E in the yeast inhibits cell growth. This growth inhibition is associated with perturbations of the actin cytoskeleton and endocytosis.
Abbreviations: T3SS, type III secretion system; T3Es, type III effectors
Keywords: AvrE, DspA/E, Erwinia amylovora, Yeast, Cellular traffic
1. Introduction
The bacterium Erwinia amylovora is the causal agent of fire blight disease of pear and apple trees. Like many other Gram-negative bacterial pathogens of plants and animals, the ability of E. amylovora to promote disease depends on a type III secretion system (T3SS) which delivers type III effector proteins (T3Es) into the host cells. The delivered T3Es act as virulence factors modulating cellular processes and suppressing host defense for the benefit of the pathogen [1–3].
DspA/E is a T3E delivered by E. amylovora which is required for disease because dspA/E mutants are non-pathogenic and unable to grow on host plants [4–6]. DspA/E belongs to the AvrE effector family of T3Es and functional cross-complementation has been demonstrated between DspA/E of E. amylovora and AvrE of Pseudomonas syringae [6]. T3Es of the AvrE family are widespread in plant-pathogenic bacteria. They are found in the genera Pseudomonas, Pantoea, Erwinia, Dickeya and Pectobacterium [7,8]. They are also found in non-pathogenic Gram-negative bacteria such as Erwinia tasmaniensis and Marinomonas mediteranneae that are associated with plants [9]. Effectors of the AvrE family are encoded by genes adjacent to the T3SS gene cluster as part of a large pathogenicity island. This suggests that they have been acquired by bacteria with the T3SS.
Effectors of the AvrE family are important to promote bacterial growth following infection. They are required for pathogenicity of E. amylovora, Pantoea stewartii subsp. stewartii and Pantoea agglomerans pv. gypsophylae [5,6,10,11] and important virulence factors for P. syringae and Pectobacterium [12,13]. Effectors of the AvrE family suppress callose deposition, a plant basal defense reaction which strengthens the plant cell wall, and also interfere with other plant defense reactions [14–16]. Furthermore, their ectopic expression in plant and yeast cells is toxic [15,17,18]. This indicates that they likely target a cellular process that is conserved between yeast and plant. Probably due to this toxicity, attempts to localize these effectors once ectopically expressed in plant cells have been unsuccessful [15,19]. Furthermore, yeast toxicity has precluded the use of the yeast two-hybrid technology to identify eukaryotic interactors of the full length proteins. Finally, effectors of the AvrE family are very large proteins which lack overall sequence homology with proteins of known function and the molecular mechanism by which they carry out their functions remains unsolved.
Most of the effectors of the AvrE family contain one or several WXXXE motifs at different locations [19,20]. WXXXE motifs have been described in T3Es from human pathogens, including Escherichia coli, Shigella spp., and Salmonella spp. These effectors perturb actin cytoskeleton of the eukaryotic host cell by mimicking constitutively active Ras-like G-proteins [21]. This suggests that effectors of the AvrE family could function as Ras-like G-proteins inducing actin cytoskeleton defects. However, a clear demonstration of intracellular trafficking perturbations with these effectors is still missing. This is probably due to the fact that expression of these effectors in plant cells promotes a rapid cell death and it is therefore difficult to observe such perturbations.
Recent studies indicate that traffic in plant shares many features with the animal and yeast models [22,23]. Intracellular trafficking has been studied for decades in yeast [24] and yeast has recently emerged as a model system for the identification and functional characterization of T3Es [25–28]. Yeast therefore provides a simple experimental model to evaluate whether a T3E induces intracellular trafficking perturbations in a eukaryotic cell.
Here we studied the physiological effects of one effector of the AvrE family, DspA/E, in Saccharomyces cerevisiae. We found that as soon as dspA/E is expressed in the yeast cell, it inhibits cell growth. We further tested whether this growth inhibition was associated with alterations of intracellular trafficking and found that dspA/E expression was associated with rapid perturbations of the actin cytoskeleton and endocytosis.
2. Materials and methods
2.1. Media, bacterial and yeast strains
Bacterial strain used in this study is E. coli DH5-α. Bacterial cells were grown in Luria Broth medium supplemented if required with 100 μg/ml ampicillin. The wild-type yeast strain used for expression of dspA/E is BY4741 (Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0, hereafter designated as BY) obtained from EUROSCARF. Yeast cells were grown in selective synthetic complete medium lacking uracil to maintain the plasmid supplemented with either 2% (w/v) raffinose, or 2% (w/v) galactose (hereafter designated as SG-URA) to induce expression of dspA/E, or 2% (w/v) glucose (hereafter designated as SD-URA) to repress expression of dspA/E.
For transformation, yeast cells were grown overnight at 30 °C in YPD medium (1% (w/v) yeast extract, 1% (w/v) bactopeptone, 2% (w/v) glucose) and transformed using the lithium acetate method (Yeast Protocols Handbook, Clontech, Palo Alto, CA).
2.2. Plasmid constructions
A fill in was performed with T4 DNA polymerase on a dspA/E XbaI/SacI fragment issued from pTB4 [15]. This fragment was then cloned into the yeast centromeric URA + plasmid pCMha189 [29] previously digested with BamHI and filled with Kleenow. A GAP repair was then performed on the obtained plasmid to introduce the pGAL promoter in front of dspA/E. The GAL1 promoter was amplified by PCR using pFA6a-kanMX6-PGAL1 plasmid as template [30] using the following primers: forward primer 5′-TTTCTCAGGTATAGCATGAGGTCGCTCTTATTGACCACACCTCTACCGGCAGATCGAGC TCTAGTACGGATTAGA-3′ and reverse primer 5′-GTGTACTGCCGCCTTGTGTTCAGTTCCCAGTGATTTTAATTCCATGTTTAAACA ATCCGGGGTTTTTTCTCCT-3′. The resulting plasmid was named pSAB191. Plasmid p416GALL [31] a centromeric URA + plasmid with a pGAL promoter, was used as control vector.
2.3. Yeast assays
To measure the growth rate, yeast cells transformed with p416GALL or pSAB191 were grown overnight in selective medium with 2% (w/v) raffinose. Cultures were then diluted to an OD600 = 0.05 and grown for 21 h in SG-URA and SD-URA media. OD600 of the culture was measured regularly. To measure cell viability, aliquots were removed regularly, 10-fold serially diluted and spotted (10 μl) onto SD-URA. Plates were incubated for 2 days at 30 °C before colonies were counted. The data presented are representative of three independent experiments.
To visualize actin patches, overnight cultures were diluted to an OD600 = 0.05 in selective raffinose containing medium and grown up to an OD600 = 0.5. Induction and repression were performed for 2 h by adding respectively 2% (w/v) galactose and 2% (w/v) glucose to the growth medium. Yeast cells were processed for actin staining with rhodamine–phalloidin (Molecular Probes Inc., Eugene, OR) essentially as described in [32]. Yeast cells were washed with water and then observed by fluorescence microscopy. Only cells with small buds were counted. Cells with most actin patches concentrated in the small bud and with 5 or fewer patches in the mother cell were classified as polarized cells. Cells with more actin patches in the mother cell than in the small bud were classified as depolarized cells. A total of 100 cells were counted during each experiment, and the data of three independent experiments were analyzed.
To analyse endocytosis, we used the fluorescent vital dye FM4-64 N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl) pyridinium dibromide, Molecular Probes Inc., Eugene, OR). For this purpose, yeast cells transformed with pSAB191 or with p416GALL were grown to the exponential growth phase with 2% (w/v) raffinose as a carbon source. Induction or repression of dspA/E were then respectively performed by adding 2% (w/v) galactose or 2% (w/v) glucose to the growth medium and cells were grown for further 2 h. The endocytosis assay using the FM4-64 was performed as described in Ref. [33]. Briefly, cells were incubated for 15 min on ice with 15 μM FM4-64. Cells were then washed and incubated in fresh medium at 30 °C. Internalization of FM4-64 was visualized during a time course of 2 h. For each condition, at least 200 cells were counted and the data of three independent experiments were analyzed.
Fluorescence observations were performed using an Olympus microscope BY61 (Olympus, Tokyo, Japan) with a rhodamine/TRITC filter set. Images were acquired with a charge-coupled device SPOT4.05 camera and processed with ImageJ software.
2.4. Quantitation of dspA/E expression
In order to quantitate the expression of dspA/E, BY pSAB191 and BY p416GALL cells were grown to exponential phase in raffinose containing medium, then induction and repression were performed for 1 h in SD-URA and SG-URA media prior to RNA extraction. RNA preparations were treated with Rnase-free Dnase Turbo (Ambion) and cDNAs were then synthesized from 400 ng of this preparation using High Capacity cDNA Reverse Transcriptase (Applied Biosystems) following the manufacturer’s instructions. Real-Time PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) following manufacturer’s instructions. A total of 10 ng of cDNAs were used to amplify dspA/E with forward primer dspA-F 5′-CGTTGCCACACCGATTAGC-3′ and reverse primer dspA-R 5′-TGGTATCTGTCCCCTCAAGGA-3′. As a reference, a constitutively expressed gene, TEF1, was amplified using forward primer TEF1-For 5′-GGCTTTCACCTTGGGTGTTA-3′ and reverse primer TEF1-Rev 5′-ATCTGGATTCGTCCCATTTG-3′. Expression of dspA/E is defined as the amount of mRNA relative to TEF1 mRNA, using the relative quantitation method. The data from three independent experiments were analyzed.
3. Results and discussion
3.1. The expression of dspA/E in yeast cells affects cell growth and is not associated with rapid cell death
To study the physiological effects induced by the expression of dspA/E in S. cerevisiae, the dspA/E gene was placed under the control of the pGAL promoter on a centromeric plasmid. The resulting plasmid named pSAB191 was introduced into the wild-type BY strain. The empty plasmid p416GALL [31] was used as a control and was also introduced into the BY strain. Yeast cultures were serially diluted and plated onto SD-URA and SG-URA solid media. On SD-URA medium, two days after plating, there was a slight difference in the size of the colonies between the strain bearing the empty plasmid and the strain bearing the pSAB191 plasmid (Fig. 1A). On SG-URA medium no growth was observed after expression of dspA/E while cells transformed with the empty plasmid were growing. This indicates that, as already observed by Oh and associates [18], the expression of dspA/E is toxic in yeast (Fig. 1A). To gain further insight into the toxicity of DspA/E, both strains were grown overnight in raffinose containing medium. The yeast cultures were then diluted to an OD600 = 0.05 in liquid SD-URA and SG-URA media and we scored the growth of the cells for 21 h (Fig. 1B). The strain bearing the empty plasmid grew faster than the strain bearing the pSAB191 plasmid in SD-URA medium. However, expression of dspA/E was not detected by RT-PCR on this media (Fig. 1C). The slight growth delay observed suggests nevertheless that an undetectable leakage expression occurs. In SG-URA medium the cells expressing dspA/E were unable to grow. This correlates with expression of dspA/E as detected by RT-PCR (Fig. 1C) confirming that the expression of dspA/E is toxic. Attempts to detect the DspA/E protein by western blot or to visualize a toxic GFP-DspA/E fusion protein after their expression in a SG-URA medium remain unsuccessful (data not shown). In the same way, attempts to detect DspA/E or WtsE, another effector of the AvrE family, once ectopically expressed in plant cells have been unsuccessful [15,19]. This suggests that proteins of this effector family are rapidly degraded following their expression in a eukaryotic cell.
Fig. 1.
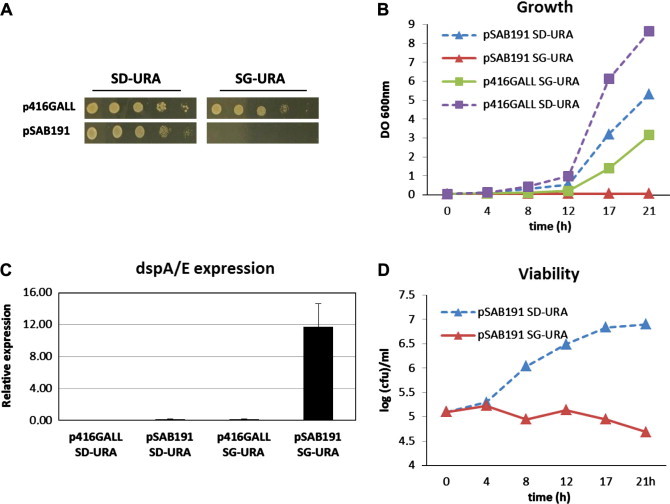
The expression of dspA/E in yeast is associated with a rapid growth inhibition. (A) Yeast cells transformed with an empty vector (p416GALL) or a plasmid bearing dspA/E under the control of a galactose promoter (pSAB191) were subjected to serial 10-fold dilution and spotted on solid SD-URA and SG-URA media. Photographs were taken after 2 days. (B) The same cells as in A were grown in SD-URA and SG-URA liquid media for 21 h and OD600 was measured at the indicated time points. (C) The same cells as in A were grown in SD-URA and SG-URA liquid media for 1 h and the expression of dspA/E was detected by RT-PCR, bars represent the expression of dspA/E. (D) Yeast cells transformed with the dspA/E expressing plasmid (pSAB191) were grown in SD-URA and SG-URA liquid media and viable cells were counted at the indicated time points.
It was previously described that DspA/E causes cell death in yeast [18] while WtsE was found to inhibit yeast growth [17]. We therefore asked whether the toxicity of dspA/E was associated with cell death in our experimental conditions. To test this, aliquots of BY pSAB191 cultures grown either in SD-URA or SG-URA media were taken regularly, serially diluted and plated onto SD-URA medium to score for viable cells (Fig. 1D). When BY pSAB191 was initially grown on SD-URA medium, the number of cells rises over time. This was expected since BY pSAB191 is able to grow in this medium (Fig. 1B). When BY pSAB191 was initially grown on SG-URA medium, the number of cells remained stable, decreasing only slightly over time (Fig. 1D). This indicates that yeast cells were not killed rapidly following the expression of dspA/E.
Altogether, our results show that the expression of dspA/E in SG-URA medium in BY is associated with a complete growth inhibition but not with a rapid death of yeast cells. In that regard it differs from the result observed by Oh and associates who reported that yeast cells are rapidly killed following expression of dspA/E [18]. However, they worked with a different yeast strain which may explain the difference.
3.2. The expression of dspA/E in yeast cells affects the actin cytoskeleton
It has been previously suggested that members of the AvrE effector family such as DspA/E may disturb vesicular trafficking. We have thus tested whether the expression of dspA/E induced perturbations of the actin cytoskeleton (Fig. 2). Strains BY pSAB191 and BY p416GALL were grown in SD-URA and SG-URA media for 2 h before labelling of actin cytoskeleton with rhodamine–phalloidin and visualization by fluorescence microscopy. When the strain BY p416GALL was grown in SD-URA medium, we observed approximately 80% of polarized cells while the strain BY pSAB191 contained only 63% of polarized cells in this medium. This slight difference is statistically significant and it indicates that, as already observed with growth experiments, dspA/E is weakly expressed in glucose-containing medium. In SG-URA medium, 80% of the BY p416GALL cells were polarized while the majority of the BY pSAB191 cells were depolarized and only 21% were polarized. This indicates that the expression of dspA/E induces actin cytoskeleton polarization defects. Such a defect can already be observed after a short period of galactose induction (1 h) (data not shown).
Fig. 2.
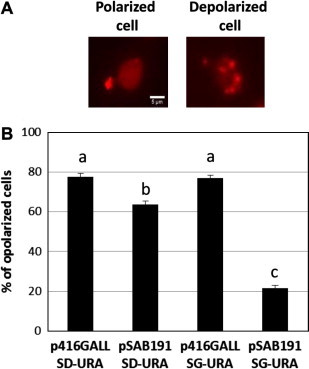
The expression of dspA/E in yeast is associated with actin cytoskeleton polarization defects. Yeast cells transformed with an empty vector (p416GALL) or the plasmid expressing dspA/E under the control of the gal promoter (pSAB191) were grown SD-URA and SG-URA media for 2 h. Cells were then processed for actin staining as described in Section 2. (A) Typical view of a polarized (left) and non-polarized (right) yeast cell is presented (scale bar: 5 μm). (B) Histograms show the average percentage of polarized cell and error bars represent the standard deviation. Columns with different letters are statistically different according to Mann and Whitney tests (P value <0.1).
3.3. The expression of dspA/E in yeast cells delays endocytosis
Endocytosis in yeast depends on a functional actin cytoskeleton [34]. As the expression of dspA/E induces actin polarization defects, we therefore asked whether it could also alter endocytosis. To test this hypothesis, we monitored endocytosis using the lipophilic styryl dye FM4-64. During the time course of endocytosis, FM4-64 is incorporated into the plasma membrane and targeted to the vacuole, via endosomes, where it accumulates at the vacuolar membrane.
BY p416GALL and BY pSAB191 cells were grown in SG-URA medium for 2 h prior to incubation with FM4-64 for 15 min at 4 °C, to inhibit endocytosis. The cells were then washed with water, resuspended in fresh SG-URA medium without FM4-64 and incubated at 30 °C. The endocytosis of FM4-64 was then visualized by fluorescence microscopy during 2 h (Fig. 3A). For the control strain BY p416GALL, after 60 min of FM4-64 chase, all the FM4-64 fluorescence was present in a regular ring structure indicating that the FM4-64 reached the vacuolar membrane (white arrows Fig. 3A1). On the contrary, the fluorescence observed in the BY pSAB191 cells was mostly present in cytosolic dots, probably corresponding to endosomes, even after 90 min of FM4-64 chase (red arrows Fig. 3A1). However, after 2 h of FM4-64 chase, 16% of the BY pSAB191 cells presented a labelling of the vacuolar membrane (Fig. 3A2). These results indicate that endocytosis is strongly delayed but not totally blocked in cells expressing dspA/E. The same results were also obtained when the expression of dspA/E was induced only for 1 h in SG-URA medium indicating that the alteration of endocytosis occurred rapidly after expression of dspA/E (data not shown).
Fig. 3.
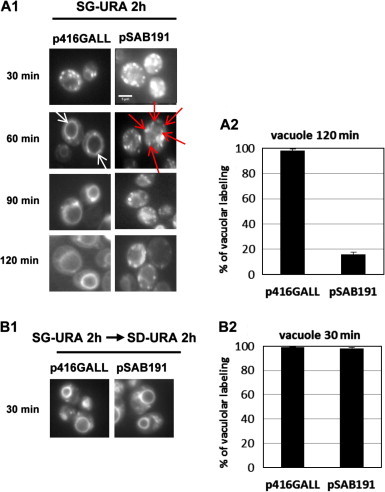
The expression of dspA/E in yeast induces endocytosis delay. (A) Yeast cells transformed with the empty vector (p416GALL) or the vector expressing dspA/E (pSAB191) were grown in SG-URA medium for 2 h prior to incubation with FM4-64. (A1) Representative pictures of the labelling obtained after 30, 60, 90 and 120 min of FM4-64 chase are presented (scale bar: 5 μM). White arrows indicate the ring structures observed when the FM4-64 is present at the vacuolar membrane. Red arrows indicate when the FM4-64 is present in dots in the cytosol (A2). Histograms show the percentage of cells presenting a labelling of the vacuolar membrane 120 min after FM4-64 chase and error bars indicate the standard deviation. (B) Yeast cells transformed with the empty vector (p416GALL) or the vector expressing dspA/E (pSAB191) were grown for 2 h in SG-URA medium and then for 2 h in SD-URA medium prior to incubation with FM4-64. (B1) Representative pictures of the labelling observed after 30 min of FM4-64 chase are presented. (B2) Histograms show the percentage of cells presenting a labelling of the vacuolar membrane 30 min after FM4-64 chase and error bars indicate the standard deviation.
We did not detect endocytosis defects when the BY pSAB191 cells were grown in SD-URA (data not shown). This indicates that endocytosis defects only occur when dspA/E is strongly expressed. This allowed us to ask whether we could reinitiate endocytosis when it has been previously blocked following expression of dspA/E. For this purpose, the cells were grown in SG-URA medium for 2 h and then, shifted in SD-URA medium for 2 h prior to FM4-64 labelling. Interestingly, 30 min after FM4-64 labelling, BY p416GALL and BY pSAB191 cells presented the same FM4-64 labelling of the vacuolar membrane (∼98% of the cells) (Fig. 3B1 and B2). This result indicates that endocytosis is rapidly restored when the expression of dspA/E is inhibited.
4. Conclusion
In the present, study we showed that expressing of the T3E DspA/E of E. amylovora in S. cerevisiae induces growth inhibition and perturbations of the actin cytoskeleton and endocytosis. To our knowledge, this is the first time these phenotypes are described for a member of the AvrE effector family. These perturbations may explain why effectors of this family suppress callose deposition at the plant cell wall [14,19].
Slight perturbations of growth rate and actin cytoskeleton polarization were observed when BY pSAB191 cells were grown in the non-inducing SD-URA medium. In this medium, we could not detect the dspA/E transcript by RT-PCR, which indicates that DspA/E is probably acting at very low concentration in the eukaryotic cells. Perturbation of endocytosis was however not sensitive to very low level of dspA/E expression as endocytosis was not altered when the cells were grown in SD-URA medium. This allowed us to test whether the DspA/E effects on endocytosis were reversible. As shifting the cells from SG-URA inducing medium to SD-URA medium clearly restores rapid endocytosis, it is likely that the targets of DspA/E are not irreversibly blocked upon DspA/E action and that the endocytosis alteration needs a sustained production of DspA/E.
In P. syringae, AvrE is functionally redundant with another T3E, HopM1. AvrE and HopM1 proteins are sequence-unrelated and the basis of this redundancy is unknown. In Arabidopsis, HopM1 targets and destabilizes an ARF-GEF protein, AtMIN7, which likely functions as a vesicle traffic regulator [35,36]. Most importantly, AvrE does not destabilize AtMIN7, the plant cell target of HopM1 [35]. Therefore, the perturbation of cellular traffic induced by members of the AvrE family is arising by a different mechanism. Although this mechanism is currently unknown, the present study highlights the fact that yeast provides a simple model system to unravel the molecular mechanism leading to these perturbations.
Acknowledgements
This work was supported by the ANR Dspcelldeath. S. Siamer was supported by an INRA “Contrat jeune scientifique”. We thank A. Thierry for providing plasmid pCMha189. We thank C. Cullin for his advices concerning the study of toxic protein in yeast and his help for the gap repair construction. We are grateful to R. Haguenauer – Tsapis for her help and support throughout this work. We also thank D. Expert for critical reading of the manuscript.
References
- 1.Block A., Li G., Fu Z.Q., Alfano J.R. Phytopathogen type III effector weaponry and their plant targets. Current Opinion in Plant Biology. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui H., Xiang T., Zhou J., Cui H.T., Xiang T.T., Zhou J.M. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cellular Microbiology. 2009;11:1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen L.-Q. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527-U199. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barny M.A., Guinebretiere M.H., Marcais B., Coissac E., Paulin J.P., Laurent J. Cloning of a large gene cluster involved in Erwinia amylovora CFBP1430 virulence. Molecular Microbiology. 1990;4:777–786. doi: 10.1111/j.1365-2958.1990.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 5.Gaudriault S., Malandrin L., Paulin J.P., Barny M.A. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Molecular Microbiology. 1997;26:1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanove A.J., Kim J.F., Wei Z.M., Kolchinsky P., Charkowski A.O., Conlin A.K., Collmer A., Beer S.V. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogdanove A.J., Bauer D.W., Beer S.V. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the hrp (type III secretion) pathway. Journal of Bacteriology. 1998;180:2244–2247. doi: 10.1128/jb.180.8.2244-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvitko B.H., Park D.H., Velasquez A.C., Wei C.-F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. Plos Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kube M. Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae. Bmc Genomics. 2010;11:11–22. doi: 10.1186/1471-2164-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frederick R.D., Ahmad M., Majerczak D.R., Arroyo-Rodriguez A.S., Manulis S., Coplin D.L. Genetic organization of the Pantoea stewartii subsp stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Molecular Plant-Microbe Interactions. 2001;14:1213–1222. doi: 10.1094/MPMI.2001.14.10.1213. [DOI] [PubMed] [Google Scholar]
- 11.Mor H., Manulis S., Zuck M., Nizan R., Coplin D.L., Barash I. Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. Gypsophilae. Molecular Plant-Microbe Interactions. 2001;14:431–436. doi: 10.1094/MPMI.2001.14.3.431. [DOI] [PubMed] [Google Scholar]
- 12.Badel J.L., Shimizu R., Oh H.S., Collmer A. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Molecular Plant-Microbe Interactions. 2006;19:99–111. doi: 10.1094/MPMI-19-0099. [DOI] [PubMed] [Google Scholar]
- 13.Holeva M.C., Bell K.S., Hyman L.J., Avrova A.O., Whisson S.C., Birch P.R.J., Toth I.K. Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Molecular Plant-Microbe Interactions. 2004;17:943–950. doi: 10.1094/MPMI.2004.17.9.943. [DOI] [PubMed] [Google Scholar]
- 14.DebRoy S., Thilmony R., Kwack Y.B., Nomura K., He S.Y. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boureau T., ElMaarouf-Bouteau H., Garnier A., Brisset M.N., Perino C., Pucheu I., Barny M.A. DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Molecular Plant-Microbe Interactions. 2006;19:16–24. doi: 10.1094/MPMI-19-0016. [DOI] [PubMed] [Google Scholar]
- 16.Degrave A., Fagard M., Perino C., Brisset M.N., Gaubert S., Laroche S., Patrit O., Barny M.A. Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Molecular Plant-Microbe Interactions. 2008;21:1076–1086. doi: 10.1094/MPMI-21-8-1076. [DOI] [PubMed] [Google Scholar]
- 17.Ham J.H., Majerczak D., Ewert S., Sreerekha M.-V., Mackey D., Coplin D. WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp stewartii, causes cell death in non-host plants. Molecular Plant Pathology. 2008;9:633–643. doi: 10.1111/j.1364-3703.2008.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh C.-S., Martin G.B., Beer S.V. DspA/E, a type III effector of Erwinia amylovora, is required for early rapid growth in Nicotiana benthamiana and causes NbSGT1-dependent cell death. Molecular Plant Pathology. 2007;8:255–265. doi: 10.1111/j.1364-3703.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 19.Ham J.H., Majerczak D.R., Nomura K., Mecey C., Uribe F., He S.-Y., Mackey D., Coplin D.L. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE Require WxxxE Motifs. Molecular Plant-Microbe Interactions. 2009;22:703–712. doi: 10.1094/MPMI-22-6-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.-S., Thammarat P., Lommel S.A., Hogan C.S., Charkowski A.O. Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. Carotovorum DspE does not suppress callose or induce expression of plant genes early in plant-microbe interactions. Molecular Plant-Microbe Interactions. 2011;24:773–786. doi: 10.1094/MPMI-06-10-0143. [DOI] [PubMed] [Google Scholar]
- 21.Alto N.M. Mimicking small G-proteins: an emerging theme from the bacterial virulence arsenal. Cellular Microbiology. 2008;10:566–575. doi: 10.1111/j.1462-5822.2007.01110.x. [DOI] [PubMed] [Google Scholar]
- 22.Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Current Opinion in Plant Biology. 2010;13:714–723. doi: 10.1016/j.pbi.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Irani N.G., Friml J. Clathrin-mediated endocytosis: the gateway into plant cells. Current Opinion in Plant Biology. 2011 doi: 10.1016/j.pbi.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 25.Siggers K.A., Lesser C.F. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host & Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curak J., Rohde J., Stagljar I. Yeast as a tool to study bacterial effectors. Current Opinion in Microbiology. 2009;12:18–23. doi: 10.1016/j.mib.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Munkvold K.R., Martin M.E., Bronstein P.A., Collmer A. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Molecular Plant-Microbe Interactions. 2008;21:490–502. doi: 10.1094/MPMI-21-4-0490. [DOI] [PubMed] [Google Scholar]
- 28.Salomon D., Dar D., Sreeramulu S., Sessa G. Expression of Xanthomonas campestris pv. Vesicatoria type III effectors in yeast affects cell growth and viability. Molecular Plant-Microbe Interactions. 2011;24:305–314. doi: 10.1094/MPMI-09-10-0196. [DOI] [PubMed] [Google Scholar]
- 29.Boyer J. Large-scale exploration of growth inhibition caused by overexpression of genomic fragments in Saccharomyces cerevisiae. Genome Biology. 2004;5 doi: 10.1186/gb-2004-5-9-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Research. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagny B., Wiederkehr A., Dumoulin P., Winsor B., Riezman H., Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. Journal of Cell Science. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 33.Vida T.A., Emr S.D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. Journal of Cell Biology. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engqvist-Goldstein A.E., Drubin D.G. Actin assembly and endocytosis: from yeast to mammals. Annual Review of Cell and Developmental Biology. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 35.Nomura K., DebRoy S., Lee Y.H., Pumplin N., Jones J., He S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 36.Nomura K., Mecey C., Lee Y.-N., Imboden L.A., Chang J.H., He S.Y. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10774–10779. doi: 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]


