FIGURE 3.
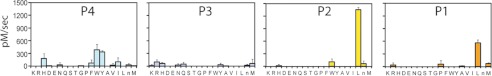
Substrate specificity of MarP determined using the complete diverse positional scanning synthetic combinatorial library. Results from P1, P2, P3, and P4 diverse positional scanning libraries are shown. The y axis represents the substrate cleavage rates in picomolar concentrations of fluorophore released per second. The x axis indicates the amino acid held constant at each position, designated by the one-letter code (with n representing norleucine). The amino acids are grouped based upon the chemical characteristics of the side chain residues (acid, basic, polar, aromatic, and aliphatic amino acids). Data are representative of three independent experiments with error bars representing ± S.D.
