Background: Initiation of protein degradation by the proteasome requires the presence of an unstructured motif, usually located at the termini of the substrate.
Results: Disruption of a hydrophobic sequence within such an unstructured motif abolishes proteasomal degradation.
Conclusion: The hydrophobicity requirement reveals a novel feature of degradation initiation sites.
Significance: Diverse proteasome substrates can be selectively stabilized by fusing a disrupted degradation initiation site to their termini.
Keywords: Endoplasmic Reticulum-associated Degradation, Proteasome, Protein Degradation, Ubiquitin, Ubiquitin Ligase, Degradation Initiation Site
Abstract
Protein elimination by the ubiquitin-proteasome system requires the presence of a cis-acting degradation signal. Efforts to discern degradation signals of misfolded proteasome substrates thus far revealed a general mechanism whereby the exposure of cryptic hydrophobic motifs provides a degradation determinant.
We have previously characterized such a determinant, employing the yeast kinetochore protein Ndc10 as a model substrate. Ndc10 is essentially a stable protein that is rapidly degraded upon exposure of a hydrophobic motif located at the C-terminal region. The degradation motif comprises two distinct and essential elements: DegA, encompassing two amphipathic helices, and DegB, a hydrophobic sequence within the loosely structured C-terminal tail of Ndc10. Here we show that the hydrophobic nature of DegB is irrelevant for the ubiquitylation of substrates containing the Ndc10 degradation motif, but is essential for proteasomal degradation. Mutant DegB, in which the hydrophobic sequence was disrupted, acted as a dominant degradation inhibitory element when expressed at the C-terminal regions of ubiquitin-dependent and -independent substrates of the 26S proteasome. This mutant stabilized substrates in both yeast and mammalian cells, indicative of a modular recognition moiety. The dominant function of the mutant DegB provides a powerful experimental tool for evaluating the physiological implications of stabilization of specific proteasome substrates in intact cells and for studying the associated pathological effects.
Introduction
Degradation signals, or degrons, are usually defined as minimal elements within proteins that are essential for the degradation of substrates by a proteolytic apparatus (1). An important property of autonomous degrons is that they are interchangeable; and once genetically fused to an otherwise long lived protein, they confer metabolic instability (2). Uncontrolled protein degradation might confer deleterious consequences at the cellular and the whole organism levels. Consequently, degron exposure is tightly regulated (2).
Protein degradation by the ubiquitin (Ub)2-proteasome system can be classified into two major categories: regulated degradation of short lived proteins and protein quality control associated degradation (PQCD). Although the enzymatic machinery that targets protein ubiquitylation is well characterized, our knowledge of the mechanism(s) by which the ubiquitylation machinery recognizes degrons is still limited (2). Understanding these mechanisms is particularly challenging in PQCD pathways, where a diverse repertoire of substrates is ubiquitylated by a limited number of ubiquitylation E3 ligase complexes, thus necessitating recognition of multiple degrons by a single E3 ligase complex.
Studies of PQCD degrons have thus far revealed a general paradigm whereby exposure of hydrophobic sequences, generally buried within the protein core, provides a degradation signal (3–7). We have previously characterized a PQCD degron in detail, using the yeast kinetochore protein Ndc10 as a model substrate (5). Ndc10, normally a stable protein, is rapidly degraded upon exposure of hydrophobic motifs localized to its C-terminal region. The degradation is dependent on the endoplasmic reticulum membrane-embedded Ub E3 ligase, Doa10. Notably, placing the degron at the C terminus of otherwise stable proteins triggers Doa10-dependent protein degradation, demonstrating its autonomous nature.
Further characterization of the Ndc10 degron revealed that it comprises two distinct and essential elements: DegA, consisting of a hydrophobic surface buried between two amphipathic helices, and DegB, a hydrophobic sequence confined to the Ndc10 loosely structured C-terminal tail (Fig. 1A). Consistent with the general PQCD degron paradigm, exposure of an intact amphipathic helix within DegA is obligatory for ubiquitylation (5). The underlying role of DegB in protein degradation has remained obscure.
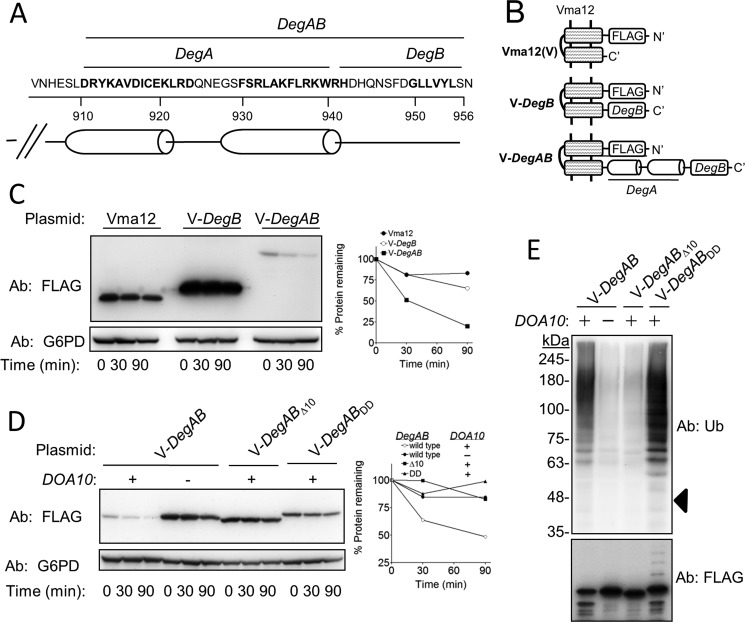
FIGURE 1.
The hydrophobic core of DegB of the Ndc10 degron is dispensable for ubiquitylation but essential for proteasomal degradation. A, amino acid sequence and structure prediction of the Ndc10 degron are shown. B, schematic presentation of Vma12-degron fusions used in C–E. V-DegB: Vma12-DegB; V-DegAB: Vma12-DegAB. C, DegB is insufficient to confer degradation to Vma12. Cells expressing the indicated proteins were subjected to cycloheximide-chase degradation assay, as described under “Experimental Procedures.” D, intact DegB is required for the degradation of Vma12-DegAB. Wild type and doa10Δ cells, expressing intact Vma12-DegAB or a truncated protein in which the last 10 aa were removed (Δ10), or a mutant protein in which two Leu residues at the hydrophobic core of DegB were replaced with Asp (DD), were subjected to cycloheximide degradation assay. E, the DD mutation within DegB did not impair the ubiquitylation of Vma12-DegAB. The various Vma12-DegAB proteins described in D were purified from cells using anti FLAG-agarose affinity gel. PolyUb chains conjugated to V12-DegAB were detected after separation on 5–15% gradient SDS-PAGE. Triangle indicates the estimated molecular mass of unconjugated V-DegAB.
We hereby report a novel role for DegB as a degradation initiation determinant. We found that in contrast to DegA, the hydrophobicity of DegB was irrelevant for ubiquitylation but was essential for degradation by the 26S proteasome. Moreover, a mutant DegB, in which the hydrophobic sequence was disrupted, acted as a dominant degradation inhibitory element when expressed at the C terminus of Ub-dependent and -independent substrates. Importantly, the inhibition of degradation by mutant DegB was not specific to yeast proteasomes, as it similarly stabilized the transcription factor XBP1, an established proteasome substrate (8, 9) expressed in the human embryonic kidney (HEK) 293T cell line.
EXPERIMENTAL PROCEDURES
Plasmids and Yeast Strains
Details are provided in Tables 1 and 2.
TABLE 1.
Yeast strains
| Yeast | Genotype | Source |
|---|---|---|
| TRy108 | MATα his3-Δ200, leu2–3112, ura3–52, lys2–801, trp1–1, gal2 | Ref. 32 |
| TRy171 | MATα his3-Δ200 leu2–3112, ura3–52, lys2–801, trp1–1, gal2, doa10-Δ1::HIS3 | Ref. 33 |
| TRy647 | MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rpn10Δ::kanMX | Open Biosystem |
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Open Biosystem |
TABLE 2.
Plasmids
| Plasmid | Relevant markers | Source |
|---|---|---|
| pTR425 | pFa6a-GFP-KanMX6 | Ref. 34 |
| pTR809 | pOC9 | Ref. 6 |
| pTR883 | pRS414 GPDp-FLAG-Vma12–6HIS-DegABΔ10-CyC1t | This work |
| pTR891 | pRS416 GPDp-Rpn10-GFP-CyC1t | This work |
| pTR892 | pRS416 GPDp-Rpn10-GFP-cODC (C441A)-CyC1t | This work |
| pTR990 | Ubiquitin CUP1 promoter (CEN/LYS2) | M. Hochstrasser |
| pTR913 | pRS414 GPDp-FLAG-Vma12-His6-DegAB-Cyc1t | This work |
| pTR1075 | pRS416 GPDp-Rpn10-GFP-DegB-CyC1t | This work |
| pTR1085 | pRS416 GPDp-Rpn10-GFP-DegBDD-CyC1t | This work |
| pTR1114 | pRS414 GPDp-FLAG-Vma12-His6-DegABDD-Cyc1t | This work |
| pTR1155 | pEFIRES-P FLAG-ATZ-mODC | Ref. 35 |
| pTR1157 | pRS414 GPD-FLAG-mODC | This work |
| pTR1266 | YCplac111 RFP-PUS1 | M. Hochstrasser |
| pTR1274 | pRS416 GPDp-Rpn10-GFP-DegBDD-CyC1t | This work |
| pTR1291 | pRS414 GPD-FLAG-mODC (Δ37)-DegB | This work |
| pTR1316 | pUC57-DegAB-DegBDD | This work |
| pTR1317 | pUC57-DegABDD-DegB | This work |
| pTR1333 | pRS414 GPDp-FLAG-mODC (Δ15)-DegB-CyC1t | This work |
| pTR1335 | pRS414 GPDp-FLAG-mODC (Δ15)-DegBDD-CyC1t | This work |
| pTR1338 | pRS414 GPDp-FLAG-Vma12-His6-DegABDD-DegB-CyC1t | This work |
| pTR1339 | pRS414 GPDp-FLAG-Vma12-His6-DegAB-DegBDD-CyC1t | This work |
| pTR 1370 | pEFIRES-P FLAG-ATZ-mODC | This work |
| pTR 1371 | pEFIRES-P FLAG-ATZ-mODC (Δ37)-DegB | This work |
| pTR1372 | pEFIRES-P FLAG-ATZ-mODC (Δ15)-DegBDD | This work |
| pTR1373 | pEFIRES-P FLAG-ATZ-mODC (Δ15)-DegB | This work |
| BT1 | pcDNA3-XBP1s | Ref. 9 |
| BT2 | pcDNA3-XBP1u | Ref. 9 |
| BT3 | pcDNA3-XBP1s-DegBDD | This work |
| BT4 | pcDNA3-XBP1u-DegBDD | This work |
Yeast, HEK293T Cells, and Bacterial Media
Yeast-rich media, minimal media, and bacterial media were prepared according to standard protocols. HEK293T cells were maintained in DMEM (10% FCS, 2 mm glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin).
Site-directed Mutagenesis
Site-directed mutagenesis was performed using Pfu Ultra polymerase, according to the manufacturer's instructions (Stratagene). All products were verified by sequencing.
Plasmid Construction
C-terminal Tagging of Mouse Ornithine Decarboxylase (mODC) and RPN10-GFP with DegB
DNA fragments containing the last C-terminal 15 amino acids (aa) of Ndc10 (DegB) were amplified with 3′- and 5′-flanking sequences complemented to regions within the C terminus of the target genes RPN10-GFP and mODC. The DNA fragments were integrated into their corresponding targets, after linearization of the backbone plasmids, by yeast recombination techniques. The presence of the 15-aa insertions was verified by sequencing.
Construction of C-terminal DegAB with Tandem Repeats of DegB
The genes encoding for DegAB-DegBDD or DegABDD-DegB were synthesized de novo and cloned into pUC57 vector by GeneScript. To generate plasmids pTR1338 and pTR1339, the relevant DNA fragments were digested from pUC57 vector using the restriction enzymes AgeI and SalI and cloned into pTR913 predigested with the same restriction enzymes.
Construction of URA3-HA-GFP-DegB
To obtain plasmids containing Ura3-HA-GFP, the GFP-encoding gene fragment from plasmid pFa6a-GFP (pTR425) was PCR-amplified and cloned into the pOC9 plasmid (pTR801) at AatII and BamHI sites, creating plasmid pTR1269. The last 100 aa of Ndc10, with or without the L950D and L951D mutations, were amplified from pTR913 and pTR1114, respectively, and cloned into the pTR1269 at BamHI and SalI restriction sites to obtain pTR1270 and pTR1272, respectively.
Construction of XBP1s-DegBDD and XBP1u-DegBDD
DegBDD was fused to XBP1s or XBP1u on pcDNA3.1 plasmids by PCR through the addition of 45 bp of the DegBDD sequence to the reverse primers. The PCR products were cloned into NotI and ApaI sites of pcDNA3.1.
Determination of Canavanine Sensitivity
Yeast cells were grown to late logarithmic phase, serially diluted 10-fold, and spotted on SD-Arg-Ura plates with 1.5 mg/ml of the Arg analog l-canavanine (Can) or on SD-Ura plates and incubated at 30 °C for 5 or 2 days, respectively. Cells were spotted at 1 A600/spot and 10-fold serial dilutions thereof.
Cycloheximide-chase
Cycloheximide-chase was performed essentially as described previously (5). Briefly, logarithmically growing cells were treated with 0.5 mg/ml cycloheximide, and culture aliquots were removed at the designated time points. After incubating the cells in 0.1 m NaOH for 5 min at room temperature, cells were resuspended in SDS gel-loading buffer and heated at 95 °C for 5 min. Proteins were then separated on 5–15% SDS-PAGE followed by immunoblotting. Antibodies used for immunoblotting were anti-FLAG mAb (Sigma-Aldrich); anti-GFP mAb and anti-hemagglutinin (HA) mAb (Roche Applied Science); anti-glucose-6-phosphate dehydrogenase polyclonal antibodies (Sigma-Aldrich). Experiments were repeated two to four times and quantified using a G-Box chemi XR imaging system and GeneTools software (Syngene, UK). Representative results are shown.
Metabolic Labeling, Pulse-Chase Analysis, and Immunoprecipitation
After starvation in methionine/cysteine-free Dulbecco's modified Eagle's medium for 45 min, HEK-293T cells, transfected with plasmids pcDNA3-XBPu-DegBDD and pcDNA3-XBPs-DegBDD were metabolically labeled with 500 μCi/ml [35S]methionine/cysteine (1,200 Ci/mmol) at 37 °C for 10 min. Pulse-chase experiments, cell lysis, and immunoprecipitation was performed as described previously (9). Purified proteins were analyzed by SDS-PAGE followed by film exposure. The experiment was repeated twice, and densitometry was performed using GeneTools software. Antibodies against XBP1 were from Santa Cruz Biotechnology. Representative results are shown.
Ubiquitylation Assay
Ubiquitylation assays were performed according to Loayza and Michaelis (10). Cells co-expressing the indicated FLAG-Vma12-DegAB plasmids, together with a plasmid containing copper-induced Ub, were incubated in selective media containing 100 μm CuSO4 until late logarithmic phase. Approximately 25 A600 cell units were harvested and then lysed by addition of 1.5 ml of 2 n NaOH, 1 m β-mercaptoethanol. The lysate was incubated on ice with 5% trichloroacetic acid. Proteins were separated by centrifugation at 17,000 × g for 10 min at 4 °C, and the pellet was resuspended in 100 μl of sample buffer. Cell extracts were diluted 30-fold with buffer supplemented with protease inhibitors (Sigma) and 5 mm N-ethylmaleimide. Extracted proteins were pulled down by incubation with anti-FLAG M2 affinity gel (Sigma) at 4 °C for 3 h. Bead complexes were washed three times and then released from the gel by incubation with 3×FLAG peptides at 4 °C for several hours while shaking. Eluted proteins were separated by 5–15% SDS-polyacrylamide gradient gel and visualized by immunoblotting, using anti-FLAG and anti-Ub antibodies.
RESULTS
The Hydrophobic Core of DegB of the Ndc10 Degron Is Dispensable for Ubiquitylation but Essential for Proteasome-mediated Degradation
To investigate the precise role of DegB in protein degradation, we constructed reporter substrates composed of the stable endoplasmic reticulum protein Vma12 (11), fused to either DegB or to DegAB which accounts for the full-length of the Ndc10 degron (Fig. 1, A and B). Cycloheximide-chase degradation experiments in yeast revealed that Vma12-DegB remained stable, whereas Vma12-DegAB was rapidly degraded in a DOA10-dependent manner (Fig. 1, C and D). Thus, DegB is essential but insufficient for triggering Vma12 degradation.
DegB comprises a hydrophobic core composed of residues Gly-Leu-Leu-Val-Tyr-Leu (Fig. 1A). To examine whether the hydrophobicity of DegB is required for degradation, we compared the stability of Vma12-DegAB with that of Vma12-DegABΔ10, where the last 10 residues of Ndc10 were deleted, and with that of Vma12-DegABDD, in which the two Leu residues at positions 950 and 951 were substituted with negatively charged Asp residues, thereby disrupting the hydrophobic core. As shown in Fig. 1D, both the truncation and missense mutations substantially stabilized Vma12-DegAB, further emphasizing the fundamental role of the intact DegB in Vma12 degradation.
To examine the role of DegB in Ub conjugation, Vma12-DegAB fusion proteins were isolated by immunoprecipitation from the respective cell extracts using anti-FLAG Abs. PolyUb conjugates in the immune complexes were subsequently detected by immunoblotting analysis with anti-Ub. As anticipated, high molecular mass polyUb conjugates were observed in wild type cells but were hardly detected in doa10Δ cells (Fig. 1E). Vma12-DegABΔ10 ubiquitylation was also barely visible, demonstrating the importance of intact DegB for ubiquitylation. Unexpectedly, ubiquitylation was substantially augmented in Vma12-DegABDD, suggesting that the hydrophobicity of DegB is required for degradation, but irrelevant for ubiquitylation. We therefore hypothesized that the hydrophobic segment is required for processes downstream to ubiquitylation.
DegBDD Abolishes Degradation if Positioned at the C-terminal Region of a Proteasome Substrate
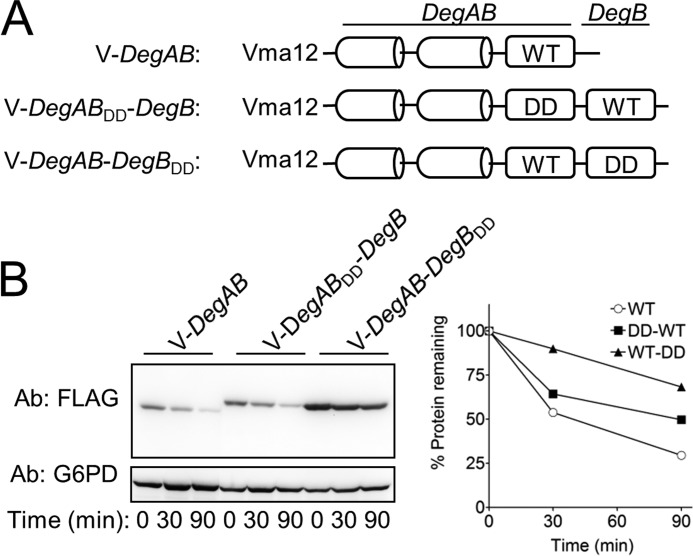
Because DegB is localized at the C terminus of Ndc10, we next investigated whether its localization is a critical determinant for degradation. To this end, we generated recombinant proteins, expressing wild type and mutant DegAB and DegB in tandem. As shown in Fig. 2B, wild type DegB restored protein degradation when fused C-terminally to the mutant degron (DegABDD-DegB), as also indicated by the similarity to the degradation kinetics of the wild type Vma12-DegAB. Thus, once proteolysis was initiated, the presence of DegBDD per se did not impede degradation. In the reciprocal experiment, when the mutant DegBDD was fused C-terminally to the wild type degron, protein degradation was substantially attenuated. The capacity of DegBDD to reverse the destabilizing properties of DegB when expressed C-terminally to it confirmed that the position of DegB at the extreme C terminus was an essential feature of the Ndc10 degron and that DegBDD operated as a dominant inhibitor of proteasomal degradation.
FIGURE 2.
DegBDD abolishes degradation when positioned at the C-terminal region of a proteasome substrate. A, schematic presentation of DegB tandem sequences used in B. B, cycloheximide-chase degradation analysis reveals that fusing DegB DD to Vma12-DegAB abolished degradation whereas fusing intact DegB to Vma12-DegABDD hardly affected degradation.
Impairment of Hydrophobicity Does Not Affect the Intracellular Localization of GFP-DegAB
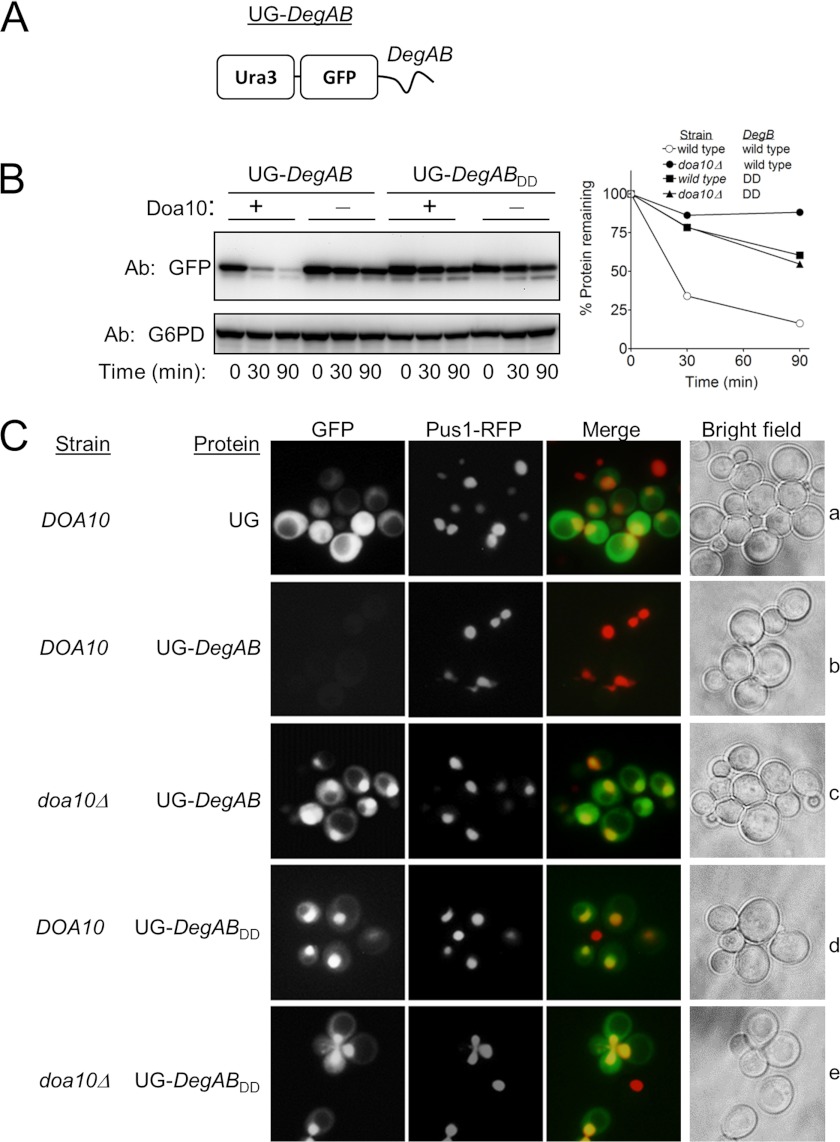
A possible explanation for the observed stabilization conferred by DegBDD was that it redirected the proteins to alternative intracellular compartments, such as inclusion bodies, where they were no longer accessible to the 26S proteasome (hence stabilized). To examine whether DegBDD influenced the cellular distribution of proteins, a reporter was constructed where the URA3 gene product orotidine-5′-phosphate decarboxylase was fused to GFP, followed by DegAB to form Ura3-GFP-DegAB (Fig. 3A). DegAB positioned C-terminally to Ura3-GFP induced Doa10-dependent degradation of the fusion protein, whereas the addition of DegABDD did not (Fig. 3B). Next, the cellular distribution of the fusion proteins in wild type and in doa10Δ cells was determined by fluorescence microscopy and compared with that of the nuclear-localized red fluorescence protein (RFP)-Pus1(12). Notably, whereas Ura3-GFP was localized in the cytosol, Ura3-GFP-DegAB, expressed in doa10Δ cells, was found mainly in the nucleus, suggesting that DegAB contained a nuclear localization signal (Fig. 3C, c and e). Ura3-GFP-DegABDD was similarly localized in the nucleus in both wild type and doa10Δ cells (Fig. 3C, d and e). Taken together, these findings indicate that substrate stabilization via DegBDD was not due to altered intracellular localization.
FIGURE 3.
DegBDD did not alter the nuclear localization of GFP-DegAB. A, schematic presentation of Ura3-GFP-DegAB (UG-DegAB). B, degradation kinetics of UG-DegAB and UG-DegABDD in wild type and doa10Δ cells are shown. Proteins levels were measured using anti GFP antibodies. C, the cellular localization of the GFP fusion proteins was determined by fluorescence microscopy. Nuclear staining of the accumulated proteins was indicated by their co-localization with Pus1-RFP, a nuclear protein. No changes in the cellular localization of the various stabilized proteins were indicated.
DegB Degron Function Is Independent of Ubiquitylation
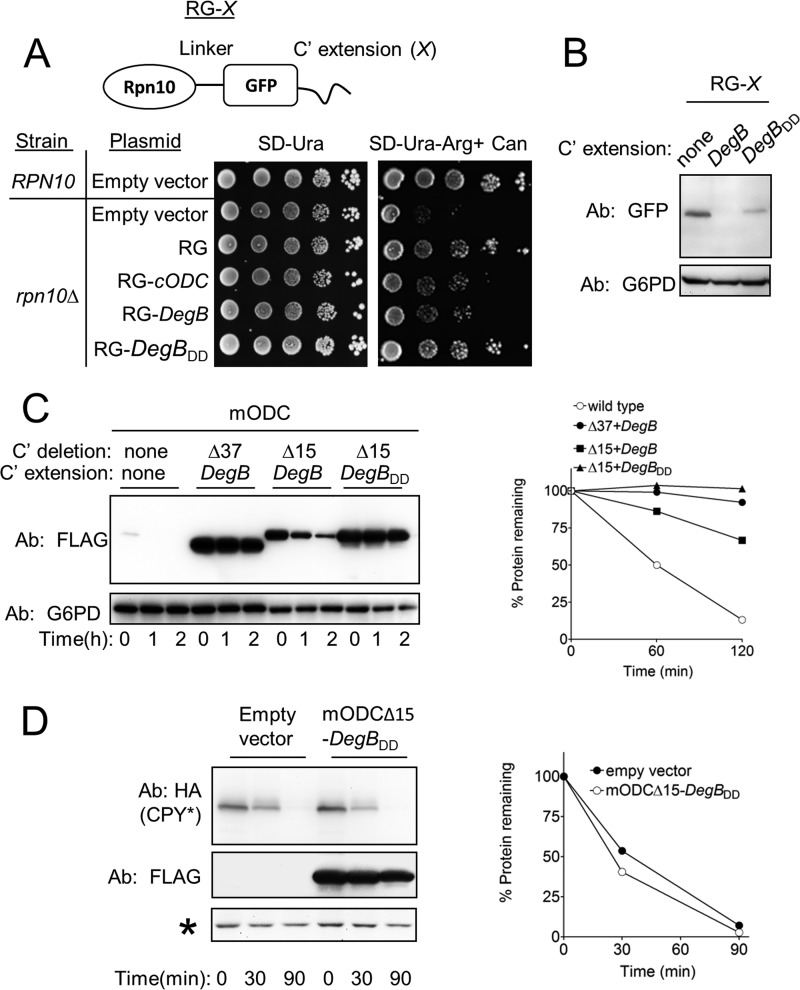
Degrons are generally defined as structural features within proteins that determine ubiquitylation, thus serving as recognition signals for E3 Ub ligases (2). We were intrigued by the finding that the disruption of DegB impaired degradation rather than ubiquitylation and postulated that DegB functions as an authentic degradation initiation site and thus, may also influence the degradation of Ub-independent 26S proteasome substrates. To test this hypothesis, we employed a yeast reporter system for Ub-independent proteasomal degradation that relies on the capacity of cells to sustain growth in the presence of Can (13), a property that is strictly dependent on active 26S proteasomes. Thus, yeast cells lacking Rpn10, an integral component of the 19S regulatory complex, are viable, but grow poorly on plates containing Can (14, 15). Expression of Rpn10-GFP that integrates into the 26S proteasome in rpn10Δ cells, reestablishes Can resistance (13). Using Can resistance as a measure of Rpn10 stability it was found that fusion of Rpn10-GFP to a C-terminal unstructured sequence, such as the 37-residue-long C terminus of ODC (cODC), destabilized Rpn10-GFP, and consequently decreased Can resistance (13). It is therefore postulated that cODC functions as a degradation initiation cue (13).
To test whether DegB can functionally substitute for cODC, the stability of Rpn10-GFP proteins, fused to various degrons, was similarly investigated by means of their ability to confer Can resistance to rpn10Δ cells (Fig. 4A). In agreement with previous findings (13), growth of rpn10Δ cells was considerably reduced in the presence of Can and was restored upon the expression of Rpn10-GFP, but not of Rpn10-GFP-cODC. Consistent with results of the Vma12-DegB stability experiments, the growth of rpn10Δ cells in the presence of Can was facilitated by expression of Rpn10-GFP-DegBDD but not by expression of Rpn10-GFP-DegB, likely indicating that the reporter protein carrying the mutant DegB was stable. This possibility was confirmed by measurement of Rpn10-GFP derivatives. Both Rpn10-GFP and Rpn10-GFP-DegBDD were readily detected whereas Rpn10-GFP-DegB was undetectable (Fig. 4B). Taken together, we concluded that DegB functioned as a degradation initiation determinant in the context of the Rpn10-GFP reporter and that this feature was absent in DegBDD.
FIGURE 4.
DegB degron function is independent of ubiquitylation. A, upper panel, schematic presentation of Rpn10-GFP fusion (RG-X), where X- is the various C-terminal extensions. A, lower panel, growth sensitivity to amino acid analog of rpn10Δ cells strains expressing RG-X is determined. Cells growing to late logarithmic phase were spotted as 10-fold serial dilutions onto SD-Ura or onto SD-Ura-Arg medium containing 1.5 μg/ml Can. Colony growth was determined after incubation at 30 °C for 4 days. B, disruption of the hydrophobicity of DegB increased the steady-state levels of RG-DegB fusion protein. C, DegBDD stabilized mODC. Degradation of mODC that was subjected to the indicated C-terminal truncations and replacements was measured in yeast by cycloheximide-chase degradation assay. D, degradation of CPY* remained intact in cells co-expressing mODCΔ15-DegAB. Asterisk, a crossreacting protein that served as a loading control. CPY*, mutant carboxypeptidase Y.
Although Rpn10-GFP-DegB is likely an Ub-independent degradation substrate, Rpn10 potentially can undergo ubiquitylation when bound to the proteasome (16–18). To further validate the Ub-independent function of DegB, we next examined its function in an Ub-independent degradation system, utilizing ODC (19, 20), which is targeted for degradation through binding to the polyamine-induced protein, antizyme (21). In yeast, the degradation of heterologously expressed mODC by the 26S proteasome is dependent on its 37-aa cODC degron (22). Hence we tested whether DegB is capable of replacing cODC in targeting mODC for degradation.
As reported previously (23), mODC was rapidly degraded in yeast (Fig. 4C). Yet, we found that replacing the 37-aa-long cODC degron with DegB stabilized the protein, most likely due to the elimination of Cys residue at position 441, which is required for recognition by the 26S proteasome (22). Indeed, when only the extreme 15 aa of mODC were replaced with DegB, thus maintaining the essential Cys-441, the mODC-DegB protein was degraded, albeit with slower kinetics than the wild type mODC. Conversely, replacing the extreme 15 aa of mODC with DegBDD considerably attenuated degradation of the protein. These results established a critical requirement for the hydrophobic region within DegB for degradation by the 26S proteasome.
Next, we considered the possibility that fusion proteins containing DegBDD bind to the proteasome, but cannot be degraded, thereby obstructing entry of other proteasome substrates to the proteasome 20S catalytic core. The data shown in Fig. 4A argue against this mode of action because DegBDD, when fused to Rpn10-GFP, reduced Can sensitivity, indicative of increased proteasome activity. To verify that indeed the proteasome remained active when expressing DegBDD, we examined the degradation of mutant carboxypeptidase Y, a PQCD substrate of the Hrd1 pathway (24). As shown in Fig. 4D, when co-expressed with mODC-DegBDD, mutant carboxypeptidase Y was degraded with the same kinetics as in cells expressing an empty vector or mODC-DegBDD. Thus, the expression of mODC-DegBDD did not significantly inhibit the proteasome activity.
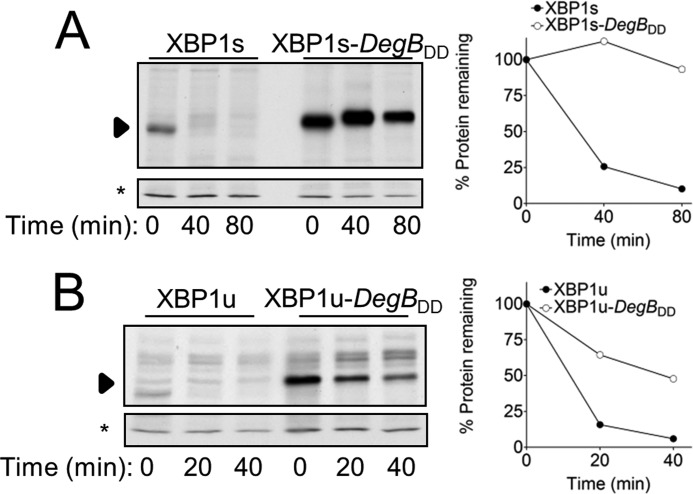
DegBDD Stabilized XBP1u and XBP1s Expressed in HEK293T Cells
We next tested whether DegBDD is also effective in stabilizing substrates of mammalian proteasomes, by fusing it to XBP1, a key transcription factor of the endoplasmic reticulum unfolded protein response. XBP1 exists in two forms: XBP1u, an unspliced form, and XBP1s, a spliced form produced upon noncanonical mRNA splicing (25, 26). XBP1s is an active transcription factor (27), whereas biological functions have not been ascribed to XBP1u, other than inhibiting XBP1s (28). Importantly, XBP1u is an extremely short lived protein that utilizes Ub-dependent and Ub-independent mechanisms of proteasomal degradation. XBP1s, on the other hand, is degraded with half-life of 30–60 min and most likely requires ubiquitylation for its destruction (8, 9). XBP1, in its two forms, thus provides an excellent model substrate to test the effect of DegBDD on proteasomal degradation in mammals. The degradation kinetics of XBP1u and XBP1s DegBDD fusion proteins were followed by pulse-chase analysis in HEK293T cells, transiently expressing the respective proteins. Similar to the results obtained in yeast, DegBDD expressed at the C terminus stabilized both XBP1u and XBP1s; albeit, the degradation of XBP1u was not completely abolished (Fig. 5B). These findings suggest a general function of DegBDD as an inhibitor of degradation of Ub-dependent and -independent proteasome substrates.
FIGURE 5.
DegBDD stabilized XBP1u and XBP1s expressed in HEK293T cells. Degradation of the indicated proteins, expressed from a pcDNA3 vector, was determined by pulse-chase analysis. Cells metabolically labeled with [35S]methionine/cysteine were sampled at the indicated time periods, followed by lysis, and immunoprecipitation, as described under “Experimental Procedures.”
DISCUSSION
In this work we have investigated the function of DegB, one of two essential elements of the Ndc10 degron. We found that an intact C-terminal DegB is required for the initiation of degradation by the 26S proteasome and that the replacement of two Leu residues with negatively charged Asp residues within the hydrophobic core of DegB inhibited proteasomal degradation of various substrates. The inhibition by mutant DegB required that it was located at the C terminus. This inhibitory function was conserved from yeast to mammals. Thus, DegBDD can operate as a universal cis-acting sequence that inhibits proteasomal degradation.
Degradation of the 26S proteasome substrates requires the presence of two essential elements: trans-acting factors that enhance substrate binding to subunits of the lid complex of the 26S proteasome and cis-elements that initiate degradation (29). The latter comprise an unstructured region that can be inserted into the catalytic cavity of the proteasome, where proteolysis occurs (13, 30). The length of the unstructured region is an important feature of the degradation initiation signal because short unstructured elements cannot trigger degradation (11). Further requirement of unstructured degradation initiation sites is a certain minimal length that separates them from the proteasome-trans-acting determinant (11).
The results presented in this study are consistent with an auxiliary role for the loosely structured DegB determinant at the Ndc10 extreme C terminus because its deletion prevented Ndc10 degradation (Fig. 1D). However, a further C-terminal truncation inhibited substrate ubiquitylation. Therefore, we have previously speculated that in the Ndc10 degron, DegB is required for both ubiquitylation and proteasomal degradation (5). Selective mutation analysis of DegB has now led us to revise this initial hypothesis and assign a degradation initiation function to DegB. This novel function is supported by the following findings: (i) disruption of the 6-aa hydrophobic core of DegB stabilized Vma12-DegAB without impairing ubiquitylation (Fig. 1E). (ii) DegB enhanced the proteolysis of Ub-independent proteasome substrates such as Rpn10-GFP (Fig. 3, A and B) and mODC (Fig. 3C) whereas DegBDD inhibited proteolysis (Figs. 1D, 2B, 3, B and C, 4B, and 5). (iii) For DegB to function, it must be localized at the C terminus where, presumably, insertion into the proteasome catalytic core of proteins containing the Ndc10 degron is initiated (Fig. 2).
As indicated above, the finding that DegBDD stabilized Vma12-DegAB without impairing ubiquitylation challenged our previous findings that removal of 10 aa from the 100-aa Ndc10 degron abolished ubiquitylation (Ref. 5 and Fig. 1E). It is possible that, unlike the selective replacement of hydrophobic aa within DegB, the removal of 10 aa disrupted the integrity of the preceding amphipathic helices within DegA and hence abolished this ubiquitylation determinant (5).
The cis-acting DegB determines degradation, provided that it is expressed at the C terminus of the protein (Fig. 2). Furthermore, when wild type and mutant DegB were expressed in tandem, the C-terminal DegB sequence element was always the dominant. This finding led us to conclude that: (i) DegB is primarily required for the initiation of proteolysis from the C terminus and (ii) DegBDD per se does not hinder degradation. It functions as a stabilizing factor only when expressed at the C terminus; but once proteolysis is initiated, DegBDD can no longer stop it. (iii) The distance between the substrate recognition motif and the initiation site is not restricted to a minimal length because extending Vma12-DegABDD by 15 aa of DegB did not alter the degradation kinetics.
It is well documented that the degradation of Ub-independent proteasome substrates such as Rpn10-GFP-cODC (Fig. 3, A and B) and mODC (Fig. 3C) requires an unstructured C-terminal region with a minimal length requirement for degradation (13, 22). Here we demonstrate that the hydrophobic aa property of the unstructured region is an additional critical feature of the degron. Our findings are in agreement with previous in vitro studies of 20S proteasome activity that indicated the existence of hydrophobic interactions between synthetic peptides and several noncatalytic proteasome sites (31). These interactions induced opening of the 20S channel via the α-rings in vitro and the release of proteolytic peptide products from the proteasome core during proteolysis (31). Notably, studies of the ATP-dependent Lon protease from Escherichia coli demonstrated a prerequisite for the presence and positioning of hydrophobic core residues within the C terminus of its substrates (32). Interestingly, Asp substitutions that disrupt hydrophobicity inhibit Lon-substrate binding and substrate degradation (31). This similarity may indicate an evolutionary conservation of elements of the proteolytic machinery, from bacteria to mammals.
Based on our findings, we propose that DegBDD can function as a dominant element for the stabilization of various short lived proteins by means of C-terminal fusion. Traditionally, to investigate the functional consequences of inhibition of proteasome-mediated degradation, proteins are stabilized either through the disruption of degrons or through the inhibition of the catalytic activity of components of the Ub conjugation machinery, mainly the proteasome. These methods are problematic in that degrons of many UPS substrates are either unknown or poorly defined and that inhibition of UPS components frequently causes pleiotropic off-target effects. The use of DegBDD offers a simple solution that circumvents these two drawbacks by enabling selective stabilization of a target protein. Our findings, that fusing DegBDD to target proteins did not affect proteasome activity (Figs. 3A and 4D), validate this experimental approach. Thus, the dominant function of mutant DegB provides a powerful experimental tool to evaluate the physiological and pathological implications of stabilizing specific proteins in intact cells.
Acknowledgment
We thank William Breuer for helpful discussions during this study and for critically reviewing the manuscript. We are grateful to P. Coffino, M. Hochstrasser, and C. Kahana for plasmids.
This work was supported by Israel Science Foundation Grant 786/08, Marie Curie International Reintegration Grant MIRG-CT-2007-205425, and the Lejwa Fund for Biochemistry (to T. R.), the Israeli Multiple Myeloma Foundation and the Israeli Cancer Association ( to B.T.).
- Ub
- ubiquitin
- aa
- amino acid(s)
- Can
- canavanine
- cODC
- C-terminal ODC
- mODC
- mouse ODC
- ODC
- ornithine decarboxylase
- polyUb
- polyubiquitin
- PQCD
- protein quality control-associated degradation.
REFERENCES
- 1. Varshavsky A. (1991) Naming a targeting signal. Cell 64, 13–15 [DOI] [PubMed] [Google Scholar]
- 2. Ravid T., Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson P. R., Swanson R., Rakhilina L., Hochstrasser M. (1998) Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94, 217–227 [DOI] [PubMed] [Google Scholar]
- 4. Fredrickson E. K., Rosenbaum J. C., Locke M. N., Milac T. I., Gardner R. G. (2011) Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol. Biol. Cell 22, 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Furth N., Gertman O., Shiber A., Alfassy O. S., Cohen I., Rosenberg M. M., Doron N. K., Friedler A., Ravid T. (2011) Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol. Biol. Cell 22, 4726–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilon T., Chomsky O., Kulka R. G. (1998) Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arteaga M. F., Wang L., Ravid T., Hochstrasser M., Canessa C. M. (2006) An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc. Natl. Acad. Sci. U.S.A. 103, 11178–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navon A., Gatushkin A., Zelcbuch L., Shteingart S., Farago M., Hadar R., Tirosh B. (2010) Direct proteasome binding and subsequent degradation of unspliced XBP-1 prevent its intracellular aggregation. FEBS Lett. 584, 67–73 [DOI] [PubMed] [Google Scholar]
- 9. Tirosh B., Iwakoshi N. N., Glimcher L. H., Ploegh H. L. (2006) Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281, 5852–5860 [DOI] [PubMed] [Google Scholar]
- 10. Loayza D., Michaelis S. (1998) Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the α-factor transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inobe T., Fishbain S., Prakash S., Matouschek A. (2011) Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 7, 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Hara L., Han G. S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeuchi J., Chen H., Coffino P. (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambertson D., Chen L., Madura K. (1999) Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lipinszki Z., Kovács L., Deák P., Udvardy A. (2012) Ubiquitylation of Drosophila p54/Rpn10/S5a regulates its interaction with the UBA-UBL polyubiquitin receptors. Biochemistry 51, 2461–2470 [DOI] [PubMed] [Google Scholar]
- 18. Kim H. T., Goldberg A. L. (2012) S5a/Rpn10, a UIM-protein, as a universal substrate for ubiquitination. Methods Mol. Biol. 832, 653–660 [DOI] [PubMed] [Google Scholar]
- 19. Kahana C., Reiss Y. (2005) Cell-free assay for ubiquitin-independent proteasomal protein degradation. Methods Mol. Biol. 301, 83–96 [DOI] [PubMed] [Google Scholar]
- 20. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 21. Murakami Y., Tanaka K., Matsufuji S., Miyazaki Y., Hayashi S. (1992) Antizyme, a protein induced by polyamines, accelerates the degradation of ornithine decarboxylase in Chinese hamster ovary cell extracts. Biochem. J. 283, 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeuchi J., Chen H., Hoyt M. A., Coffino P. (2008) Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase. Biochem. J. 410, 401–407 [DOI] [PubMed] [Google Scholar]
- 23. Mamroud-Kidron E., Kahana C. (1994) The 26S proteasome degrades mouse and yeast ornithine decarboxylase in yeast cells. FEBS Lett. 356, 162–164 [DOI] [PubMed] [Google Scholar]
- 24. Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 25. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 26. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 27. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. (2003) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 100, 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 31. Gur E., Sauer R. T. (2008) Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 22, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74, 357–369 [DOI] [PubMed] [Google Scholar]
- 33. Swanson R., Locher M., Hochstrasser M. (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 35. Bercovich Z., Kahana C. (2004) Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin-dependent and is inhibited by antizyme. J. Biol. Chem. 279, 54097–54102 [DOI] [PubMed] [Google Scholar]