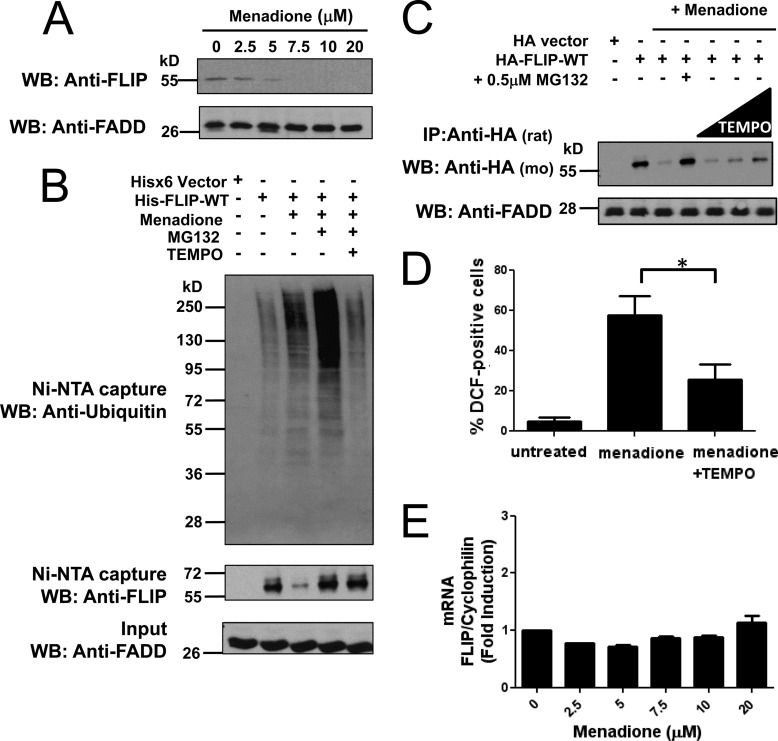
FIGURE 1.
ROS-dependent ubiquitination and degradation of FLIP. A, PPC1 cells were treated with increasing concentrations of menadione for 8 h. Total cell lysates were analyzed by immunoblotting using a specific mouse anti-FLIP antibody. FADD served as a loading control. B, PPC1 cells were transfected with pcDNA3.1-His6 vector (first lane) or plasmid encoding His-FLIP (second to fifth lanes) for 16 h. Cells were treated with menadione (5 μm) in the presence of MG132 (1 μm) or TEMPO (1 mm) as indicated for 10 h. Multiple dishes with identical treatments were pooled before lysis. His-tagged FLIP proteins were captured by Ni-NTA resin and eluted by imidazole solution. The inputs (1/20 of lysates used for FLIP protein capture) and the Ni-NTA-purified proteins were analyzed by immunoblotting using mouse anti-FLIP, anti-Ubiquitin, and rabbit anti-FADD antibodies. C, PPC1 cells were transfected with either pcDNA3-HA vector (first lane) or HA-tagged FLIP-WT plasmid (second to seventh lanes) for 16 h. Cells were then treated with menadione (5 μm) in the presence of MG132 (0.5 μm) or increasing concentrations of TEMPO (500 μm, 1 mm, and 2 mm) for 8 h. Cell lysates were immunoprecipitated (IP) using rat anti-HA antibody and the immunoprecipitated proteins were analyzed by immunoblotting using mouse anti-HA to detect FLIP. The inputs (1/10 of lysates used for immunoprecipitation) were analyzed by immunoblotting using rabbit anti-FADD as loading control. D, PPC1 cells were treated with menadione (30 μm) with or without TEMPO (1 mm) for 8 h. The adherent cells were incubated with H2DCFDA for 30 min at 37 °C, washed, collected, and analyzed by FACS. Statistical significance (mean ± S.E.; n = 4) was determined by one-way analysis of variance and Tukey post-test. *, indicates the p value is p < 0.05. E, PPC1 cells were treated with increasing concentrations of menadione for 8 h and then total RNA was extracted from cells. Relative levels of endogenous FLIP mRNA were assessed by quantitative RT-PCR.