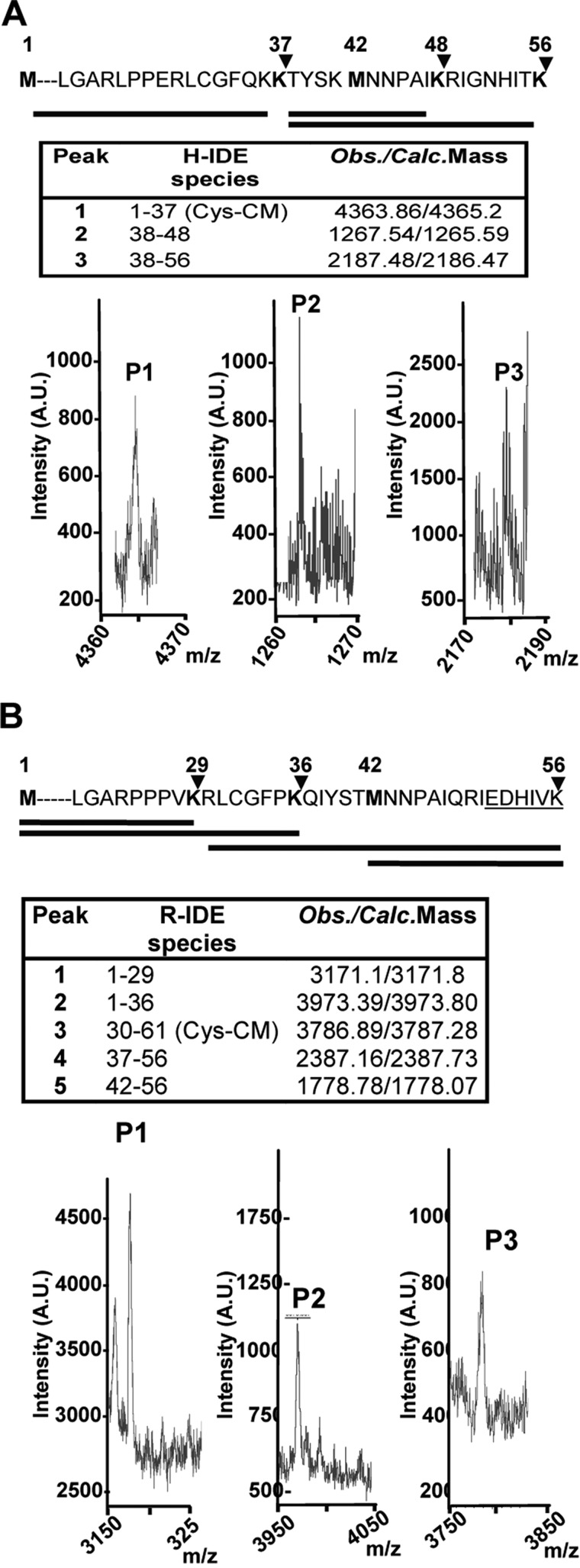
FIGURE 2.
In vivo identification of IDE-Met1 isoform. A and B, upper panels, schematic representation of the amino acid sequences (1–56) of hIDE (A) and rat IDE (B) showing two alternative in-frame initiation Met residues, Met1 and Met42 (bold). The arrowheads indicate the predictive cleavage sites in the N terminus of IDE-Met1 isoform by endo-Lys-C. Solid lines below depict the expected peptides obtained after digestion. A and B, lower panels, MALDI-TOF MS analysis of the peptides (P1, P2, and P3) obtained after endo-Lys-C digestion. Insets, hIDE (H-IDE) and rat IDE (R-IDE) species and expected/calculated masses of predicted peptides. Cys-CM, carboxymethyl-Cys; A.U., arbitrary units; Obs., observed; Calc., calculated.