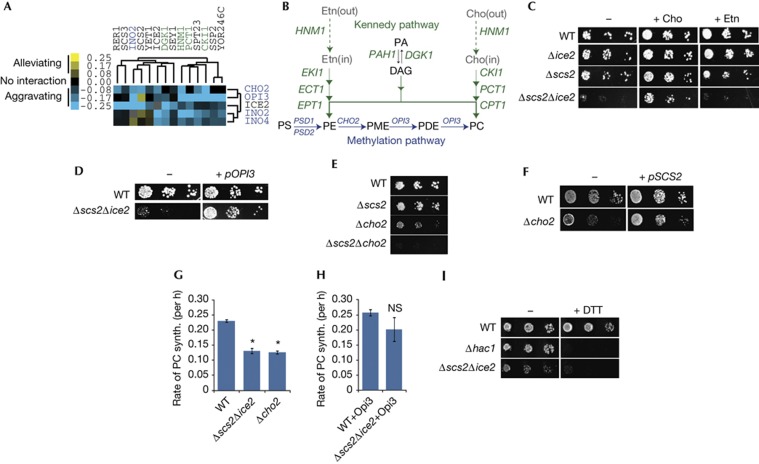
Figure 1.
Opi3 function is compromised in Δscs2Δice2 cells. (A) Gene clusters containing SCS2 and ICE2 revealed a potential role for PM–ER contacts in phospholipid metabolism. Genes are colour-coded according to pathways in (B). Data were clustered from Costanzo et al [12]. (B) Principal pathways of phospholipid synthesis in S. cerevisiae. Genes encoding enzymes required for each step are shown in italics. (C) Ten-fold serial dilutions of the indicated mutant yeast strains grown on SD media (−) or SD supplemented with either +Cho or +Etn. (D) WT and Δscs2Δice2 yeast grown on SD media transformed with either a plasmid expressing Opi3 (+ pOPI3) or a control plasmid (−). (E) Mutant yeast grown on SD media without choline. (F) Yeast grown on SD media transformed with either a plasmid expressing Scs2 (+ pSCS2) or a control plasmid (−). (G) In vivo PE methylation assay. Log phase yeast were pulse-labelled with [3H]-Etn and the rate of PC synthesis was determined by measuring conversion of PE into PC over time. Error bars, s.e.m. Asterisks, P<0.005 versus WT. (H) PE methylation assay for indicated strains expressing Opi3 from a plasmid (NS versus WT). (I) Yeast grown on SD media containing choline with 1 mM DTT (+DTT) or without (−). Cho, choline; DAG, diacylglycerol; DTT, diothiothreitol; ER, endoplasmic reticulum; Etn, ethanolamine; NS, not significant; PA, phosphatidic acid; PC, phosphatidylcholine; PDE, phosphatidyldimethylethanolamine; PE, phosphatidylethanolamine; PM, plasma membrane; PME, phosphatidylmonomethylethanolamine; PS, phosphatidylserine; SD, synthetic-defined; synth, synthesis; WT, wild-type.