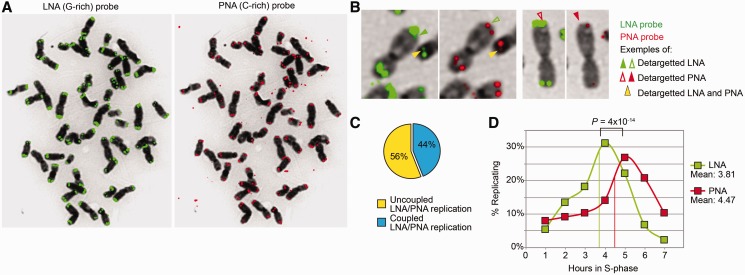
Figure 2.
Large telomere-like sequences detected by LNA telomeric probes replicate independently of bona fide gorilla telomeres. (A) G-rich LNA (left, in green) and C-rich PNA (right, in red) probes yield different telomere FISH profiles on the same gorilla metaphase. The C-rich PNA probe detects round and well-defined fluorescent signals, typical of telomeres. The G-rich LNA probe detects a much larger and not so well-defined area, which is associated with the heterochromatic caps. Contrary to the PRINS technique (29), the LNA probe does not detect paracentromeric telomere-like sequences present on particular gorilla chromosomes (29). Of note, extrachromosomal telomere signals are uniquely recognized by the C-rich probe (right). (B) The ReDFISH approach allows to examine the dynamics of replication of sequences revealed by PNA alone, LNA alone or both. The figure shows examples of extremities for which the detargeting occurred either simultaneously (yellow arrowhead), specifically for the LNA probe (green arrowhead) or specifically for the PNA probe (red arrowhead). (C) The percentage of LNA-revealed and PNA-revealed telomere sequences that replicated at different moments, thus suggesting distinct replication timings, is higher than that of sequences replicating together. (D) The replication of LNA-revealed (presumably subtelomeric) telomere sequences peaks significantly earlier than that of PNA-revealed (bona fide) telomere sequences (Fisher exact test), suggesting that the former follow a different replication program. Mean replication timings are indicated for both structures.