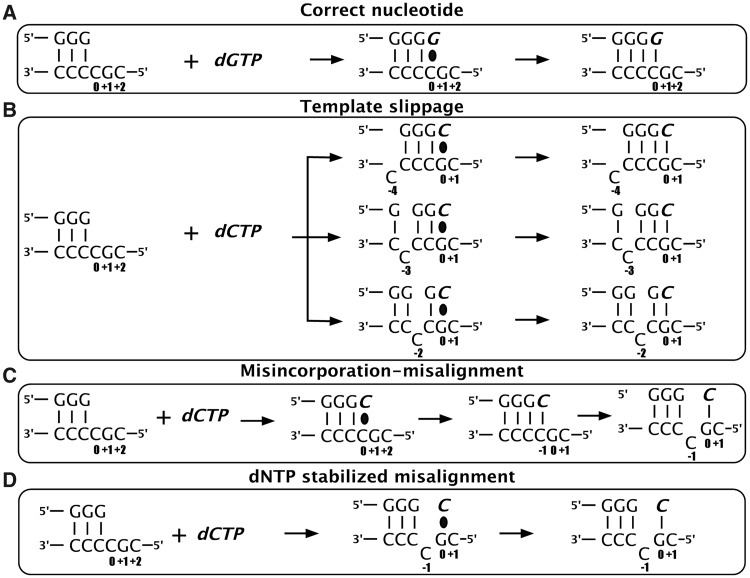
Figure 1.
Single-base deletion mechanisms. On a deletion ‘hotspot’ containing a homopolymeric run of pyrimidines followed by a 5′G, DinB polymerases could (A) add the correct nucleotide (dGTP) or generate single-base deletions in the presence of the incorrect nucleotide (dCTP) by any of three proposed mechanisms: (B) template slippage (C) misincorporation–misalignment and (D) dNTP-stabilized misalignment (see text for more details). The incoming nucleotide and newly added base are shown in italics (bold). The base positions assigned are shown with respect to the templating position defined as 0. The black oval represents base-pairing before phosphodiester bond formation.