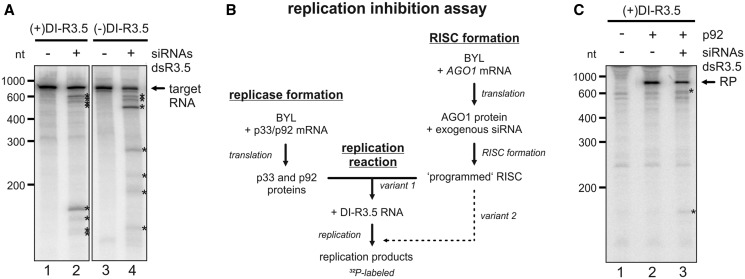
Figure 3.
Antiviral RNA silencing with a virus-derived siRNA pool. (A) ‘RISC formation/cleavage assay’ with DI-R3.5 RNA. A pool of siRNAs was generated by RNase III (ShortCut®) cleavage of dsR3.5 RNA. Using BYL where AGO1 was overexpressed by in vitro translation, RISC was formed with this siRNA pool. 32P-labeled (+) or (−)DI-R3.5 RNA transcripts were added to the extract that contained the programmed RISC, and the reaction products were subsequently analyzed by denaturing PAGE and autoradiography (lanes 2 and 4). The analogous experiments performed with a non-related (‘gf698’) siRNA served as negative controls (lanes 1 and 3). The positions of the labeled target RNAs are indicated; most prominent cleavage products are indicated by asterisks. (B) Schematic representation of the in vitro ‘replication inhibition assay’. A BYL reaction mixture that contained in vitro translated AGO1 and RISC that was ‘programmed’ with the siRNA(s) of choice was added to a second BYL reaction mixture that contained the in vitro translated TBSV replicase proteins p33 and p92. In experimental ‘variant 1’, RNA replication was initiated by combining both reactions and by the subsequent addition of replication mix and DI-R3.5 RNA template. In experimental ‘variant 2’, the mixture that contained the programmed RISC was added at a later time point to the replication reaction (Figure 4E). (C) ‘replication inhibition assay’. The reaction described in (B) was performed with (+)DI-R3.5 RNA. Lane 1; in the absence of p92 (no replication). Lane 2; with an unrelated (‘gf698’) siRNA (negative control). Lane 3; with the dsR3.5-generated vsiRNA pool. The RNA replication products (RP) are indicated, as well as the most prominent RNA cleavage products (asterisks).