Figure 1.
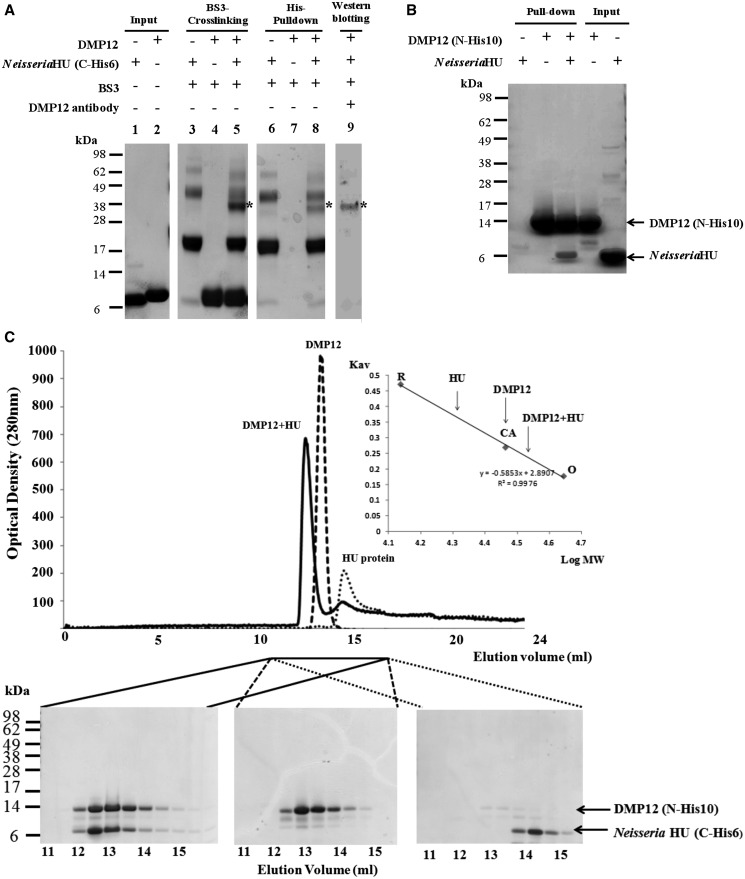
BS3 cross-linking, His–pull-down and gel filtration assays all demonstrate an interaction between Neisseria DMP12 and HU. (A) A BS3 cross-linking assay demonstrates an interaction between DMP12 and C-terminal His6-tagged Neisseria HU. After the addition of BS3, shifted bands that indicated protein–protein cross-linking could be seen (lane 5); the shifted bands are marked with an asterisk. C-terminal His6-tagged Neisseria HU was pulled down using Ni-NTA beads, and cross-linked DMP12/ C-terminal His6-tagged Neisseria HU was detected (lane 8). The presence of DMP12 in the shifted bands was confirmed by using anti-DMP12 antibody (lane 9). (B) In this His–pull-down assay, HU without any tag was used as prey, and N-terminal His10-tagged DMP12 was used as bait. HU was pulled down by His10-tagged DMP12. (C) The molecular weights of DMP12, Neisseria HU and DMP12/HU complex were measured by gel filtration on a Superose 12 HR 10/30 column monitored at 280 nm. The standard proteins ovalbumin (OA; 43 kDa), carbonic anhydrase (CA; 29 kDa) and RNase A (RA; 13.7 kDa) were fractionated on the same column and used to generate a plot of Kav against log MW (inset). The Kav of each target protein (DMP12, HU and DMP12/HU complex) was then used to find its molecular weight (The DMP12 peak seems to correspond to its dimeric form in the low ionic buffer (20 mM Tris, pH 7.4, and 5 mM MgCl2) that was used in this gel filtration. However, in a higher ionic strength buffer, such as PBS (pH7.4), DMP12 only appears in its monomeric form; see Table 2, footnote C).