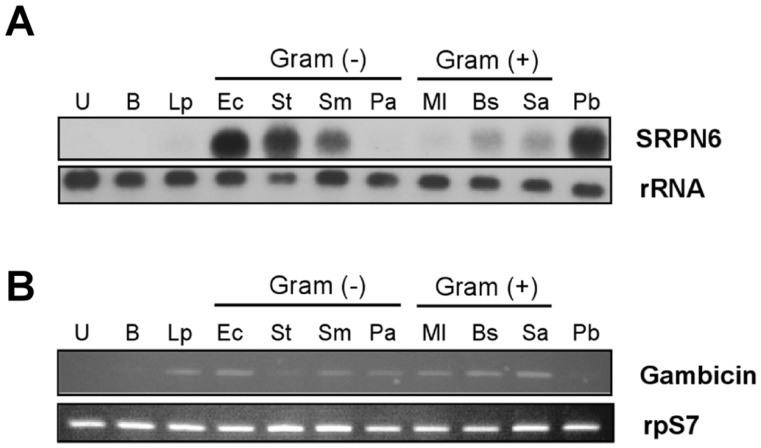
Figure 1. SRPN6 is differentially induced by bacteria in the mosquito midgut.
(A) Bacteria (1×106/ml of buffer; 2,000 bacteria assuming ingested volume of 2 µl) or the indicated component were fed to An. stephensi mosquitoes and their midguts were dissected 6 h later. Total RNA (3 µg) was analyzed by Northern blot using a 32P-labeled SRPN6 cDNA probe (upper panel). The blot was then stripped and hybridized with mitochondrial rRNA probe as a loading control (lower panel). Samples are identified above each lane as follows. U: unfed control; B: buffer-fed; Lp: E. coli LPS (10 mg/ml); Ec: Enterobacter cloacae; St: Salmonella typhimurium; Sm: Serratia marcescens; Pa: Pseudomonas aeruginosa; Ml: Micrococcus luteus; Bs: Bacillus subtilis; Sa: Staphylococcus aureus; Pb: P. berghei-infected blood, analyzed 24 h after feeding (positive control). (B) Expression of gambicin, a mosquito anti-microbial peptide. Gambicin transcript abundance was analyzed by semi-quantitative RT-PCR (upper panel) using ribosomal protein S7 (rpS7) mRNA expression as a loading control (lower panel). RNA templates are the same as those used in panel (A).