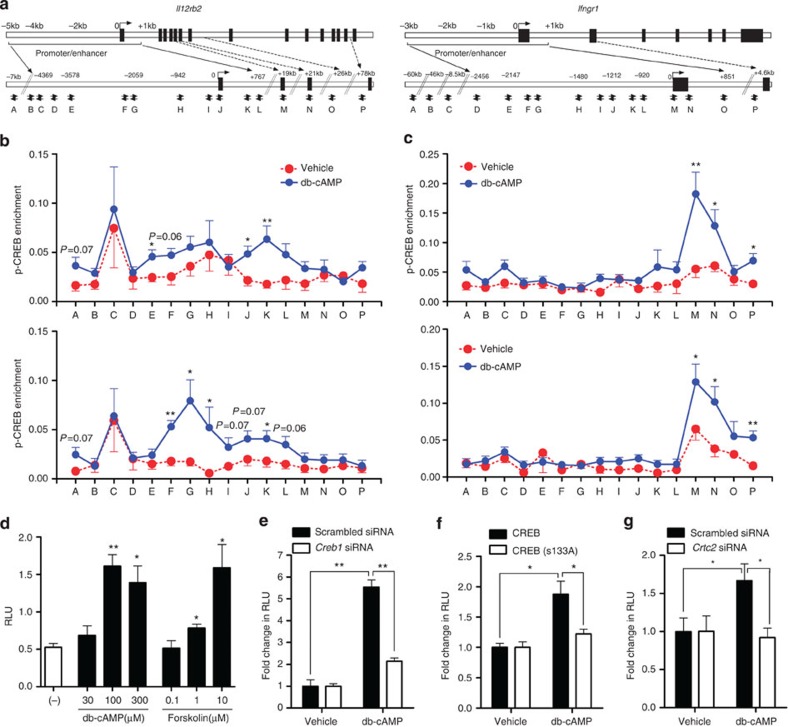
Figure 6. Recruitment of p-CREB and CRTC2 to Il12rb2 and Ifngr1 gene loci upon cAMP stimulation.
(a) Schematic diagram of the mouse Il12rb2 (left) and Ifngr1 (right) genes and their promoter/enhancer regions illustrating potential CRE sites and the positions of the primers. (b,c) Enrichment of p-CREB(S133) (upper) and CRTC2 (lower) at the Il12rb2 (b) and Ifngr1 (c) gene loci. T cells were rested for 2 days followed by stimulation with db-cAMP or vehicle for 1 h. Cells were fixed, and chromatin immunoprecipitation was performed using anti-p-CREB (S133) and anti-CRTC2. Data are normalized to the input DNA and represent eight individual chromatin immunoprecipitations from four independent experiments (mean±s.e.m.). (d–g) Luciferase activity of EL4 cells stably expressing the Il12rb2 luciferase reporter, stimulated for 24 h with indicated concentrations of db-cAMP, forskolin or vehicle (d), or stimulated for 24 h with db-cAMP after 40 h treatment with scrambled or Creb1 siRNA (e), after 24 h transfection with WT CREB or the CREB(S133A) mutant (f) or after 40 h treatment with scrambled or Crtc2 siRNA (g). Relative light unit (RLU) is normalized to vehicle control of each transfection condition (e–g). Data shown as mean±s.e.m. are representative of two (d–g) independent experiments with triplicates. Statistical significance was examined by unpaired two-tailed Student’s t-test, *P<0.05; **P<0.01.