Abstract
Purpose
To determine the prevalence of South Amerindian Y chromosome in Chilean patients with spermatogenic failure and their association with classical and/or AZFc-partial Y chromosome deletions.
Methods
We studied 400 men, 218 with secretory azo/oligozoospermia (cases) and 182 controls (116 fertile and/or normozoospermic, and 66 azoospermic with normal spermatogenesis). After a complete testicular characterization (physical evaluation, hormonal and/or biopsy) peripheral blood was drawn to obtain DNA for Y chromosome microdeletions, AZFc-partial deletions and biallelic analysis by allele specific polymerase chain reaction (PCR) of the M3 (rs3894) single nucleotide polymorphism (SNP).
Results
Classical AZF microdeletions were found in 23 cases (Y-microdeleted). AZFc-partial deletions were observed in 10 cases (6 “gr/gr”, 3 “b2/b3” and 1 “b1/b3”) and 4 controls (4 “gr/gr”). The AZFc-partial deletions were mainly associated with the absence of DAZ1/DAZ2 (64 %). No significant differences in the prevalence of AZFc-partial deletions were observed between cases and controls. We observed a significant higher proportion of the Q1a3a haplogroup in Y-microdeleted men compared to patients with spermatogenic failure without deletions and control men (P < 0.01 and P < 0.05, respectively by Bonferroni test). Among them, patients with AZFb deletions had an increased prevalence of the Q1a3a haplogroup compared to controls, cases without deletions and to those with complete or partial-AZFc deletions (P < 0.01, Bonferroni test).
Conclusions
The Q1a3a South Amerindian lineage seems to increase the susceptibility to non AZFc microdeletions. On the other hand, in Chilean population the AZFc-partial deletions (“gr/gr”, “b1/b3” and/or “b2/b3”) does not seem to predispose to severe spermatogenic impairment.
Keywords: Y chromosome microdeletions, AZFc-partial deletions, Male infertility, South Amerindian Q1a3a haplogroup
Introduction
Primary spermatogenic failure is largely responsible for male infertility, but its etiology remains unclear in nearly half of all cases [3, 17, 25]. Until now, Y chromosome microdeletions constitute the most important known etiological factor for primary spermatogenic failure, with a prevalence of approximately 15 % in subjects with azoospermia or severe oligozoospermia [15, 16, 24, 31, 37, 41]. In addition to Y chromosome microdeletions and other less frequent mutations [6, 12, 14], several studies have proposed that there may be other factors, possibly associated with the environment and/or specific genetic background, that may affect the susceptibility to suffer spermatogenic defects [1, 14, 32, 33, 39, 44, 45].
Regarding the Y chromosome, several studies have suggested that some lineages or haplogroups of the Y chromosome may confer susceptibility to complete- or partial- AZFc deletions [1, 44, 45]. There are various known binary markers whose low rate of mutation represent unique events in the evolutionary process, allowing to build hierarchical phylogenetic haplogroups of the Y chromosome [10, 23]. The Y-SNP M3 (DYS199 locus) is a single nucleotide polymorphism (SNP) whose C > T mutation define the Q3 (YCC2002)[10] or Q1a3a (YCC2008) [23] haplogroup of the Y chromosome, which is almost totally restricted to the Americas [22, 46]. To our knowledge until now, there are no studies of Y chromosome haplogroups in mixed or native South American populations with Y chromosome microdeletions.
The study of DYS199 biallelic locus in human populations has shown that T allele is found almost exclusively in South and Central Native American populations, with the prevalence in the indigenous North America population approximately 50 % [40]. Instead, the C allele has been observed in humans outside the Americas and nonhuman primates, indicating that the mutation at M3 (T allele) originated very early and represents the common Y chromosome lineage for native South America populations. In fact, the frequencies of the T allele at Y-SNP M3 differed significantly between Hispanics [18] and the native populations of South America [4, 40].
The genetic background of Chilean population is mainly formed by an asymmetrical mating between ancestral populations of Spanish conquerors and Aboriginal natives, where the prevalence of Amerindian Y chromosome based on the DYS199 and DYS19 loci is much lower (<20 %) than Amerindian autosomal markers (around 40 %) [8, 9].
The study of Zhang et al. [47] showed that partial-AZFc deletions are characteristic of men with the Q1 and N1 haplogroups, which also have an increased incidence of complete-AZFc deletions [47]. Therefore, partial-AZFc deletions can be considered as a candidate cause of increased susceptibility to complete AZFc deletions. In addition, some studies in Han Chinese have observed that genetic background of Y chromosome may affect the formation of “gr/gr” partial-AZFc deletion, and may contribute to spermatogenic impairment [44]. Even though the genetic susceptibility to spermatogenic failure can be influenced by the AZFc-partial deletions, it also has been related to other structural changes not related to these AZFc deletions [45]. In fact another study from Northern Italy observed a higher prevalence of the E haplogroup among patients with complete AZFc deletions (“b2/b4”)[1].
In this study, we investigated the proportion of Amerindian Q3-M3 haplogroup in Chilean patients with primary spermatogenic failure, with or without Y chromosome microdeletions and AZFc-partial deletions, versus demographically comparable controls without failure in spermatogenesis.
Methods
Subjects
This study was approved by the Ethical Review Board of the Central Metropolitan Health Service, Santiago, Chile, and all subjects gave their informed consent. We studied 284 selected Chilean infertile patients who consulted for infertility at the Institute of Maternal and Child Research, San Borja-Arriarán Clinical Hospital, or at the José Joaquín Aguirre Hospital, Santiago, Chile. Two fifty two of these patients were referred for testicular biopsy for diagnosis and/or spermatic recuperation by testicular sperm extraction (TESE) between March 2003 and December 2010. This procedure was performed to men with a minimum of 1 year of infertility and in whom previous semen analyses had shown azoospermia (78 %) or low numbers of viable spermatozoa with sperm count ≤5.0 × 106/mL. Thirty two of the 284 infertile patients did not undergo a testicular biopsy, but they were included because they had Y chromosome microdeletions and/or were azoospermic with a high serum FSH associated with a reduced testicular volume, measured by the Prader orquidometer (<15 cc). Patients underwent an evaluation that included a complete physical examination, hormonal studies, karyotype and Y chromosome microdeletions (complete and partial AZFc).
We selected additional 116 healthy men, of which 99 were normozoospermic and 17 have reported fertility, but without seminal analysis.
Subjects were excluded if they had hypogonadotrophic hypogonadism, hyperprolactinaemia, abnormal karyothype, chronic diseases, clinical varicocele, retractile testis, male accessory gland infections, orchitis, genital trauma, drugs consumption and concomitant hormonal treatment.
All subjects were grouped according to their place of residence in the different geographic regions of Chile (http://www.arabe.cl/chile/comunas.html) including their location within different Districts of Santiago (the capital of Country) and the Metropolitan region (RM, Region of the capital).
Hormonal measures
Serum concentrations of LH and FSH were measured by immunoradiometric assay (Siemens Medical Solutions Diagnostics, LA, CA, USA). Total testosterone was measured by radioimmunoassay (Diagnostic System Laboratories, Webster, TX, USA). Blood samples were collected between 8 and 10 AM.
Semen analysis
Semen analysis was performed according to the World Health Organization criteria [43]. The diagnosis of azoospermia was based on the absence of sperm in at least two separate semen analyses after centrifugation of semen samples (1,000 g, 5 min). Infertile patients and normozoospermic healthy volunteers underwent at least two semen analyses. Sperm morphology evaluation using the Kruger’s strict criteria [26] was also performed in normal controls.
Determination of Y chromosome microdeletions and AZFc-partial deletions
DNA samples were isolated from peripheral blood using the Wizard® genomic DNA purification kit (Promega, WI, USA). All infertile and fertile subjects were evaluated with an standard set of 20 Y-specific STS primers in 6 multiple PCR reactions (Mix1: sY85, sY83, sY90, sY221; Mix2: sY143, sY158, sY255; Mix3: sY142, BPY2, sY98; Mix7: sY153, XKRY, CDY2; Mix9: EIF1AY, USP9Y; Mix11: DBY, TSPY, F19/E355) as previously was described [7]. Further characterization of the Y-chromosome microdeleted patients were performed in single PCR reactions with the remaining 16 of 34 primers, as previously described [7] (Fig. 1). The same PCR conditions were performed for primers sY1227 and sY1228 specific to spacer to arm boundaries on palindrome P5 and sY579 specific for spacer on palindromes P1 and P2 [28, 34].
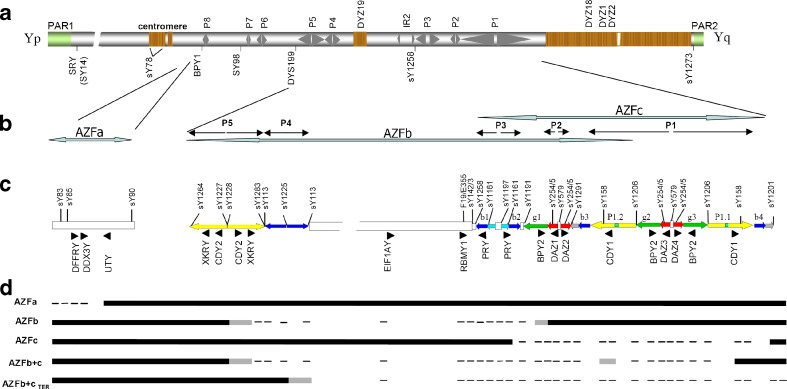
Fig. 1.
Schematic Y chromosome structure and STS employed to detect deletions. a Schematic view of the Y-chromosome with pseudoautosomal regions (PAR) 1 and 2, palindromes 1–8, centromere, heterochromatin regions, inverted repeat IR and the chromosomal position of the DYS199 locus as depicted in Lange et al. [28]. b Expanded view of AZFs regions. c The AZFb and AZFc amplicon structure is drawn according to the color code of Kuroda-Kawaguchi et al. [27]. In (a) and (c) are shown the location of the principal STS and locus analyzed for the screening of AZFs microdeletion and AZFc partial deletions of the Y chromosome. d Exemplary results of PCR-based analysis in patients with AZFa, AZFb, AZFc or AZFb+c microdeletion. Black bar: STS/locus present. Lines: STS/locus absent. Gray lines: STS/locus that normally amplifies by PCR but assumed to be absent in the context of deletion pattern
In order to detect AZFc-partial deletions, all subjects were studied through a three-step sequence-tagged site (STS) screening: the first step included sY1291 and sY1191, whose absences are specific to “gr/gr” and “b2/b3” AZFc-partial deletions respectively [38]. In case of negative amplifications, our screening included confirmation of these AZFc-partial deletions by the presence of sY1161 (i.e. “b1/b3” excluded), sY1258, sY1197, sY1206 and sY1201 [38]. The “b1/b3” AZFc-partial deletions were determined by the absence of sY1291, sY1191, sY1161 and sY1197. The third step of screening was performed to confirm the AZFc-partial deletions by the absence of DAZ1/DAZ2 or DAZ3/DAZ4, discriminating for different DAZ genes copies (DAZ2-DAZ4) by DAZ-SNVs and STS specific as previously described [29].
Genotyping of the Y-SNP M3 (DYS199 locus)
The Q1a3a lineage (YCC2008) was determined in all subjects by detection of the T allele on Y-SNP M3 (rs3894). Genomic DNA isolated from peripheral blood was used for analysis of C or T allele by allele-specific polymerase chain reaction (PCR) according to Underhill et al. [40] and Cifuentes et al. [8] with some modifications. Briefly, two polymerase chain reactions were done for each DNA sample in parallel. One reaction was performed using the C-specific reverse primer (5′-GGT ACC AGC TCT TCC TAA TTG-3′) [40], while the other was performed using the T-specific reverse primer (5′-GGT ACC AGC TCT TCC TAA TTA-3′) [40]. In both reactions, the DYS199 forward primer was the same (5′-TAA TCAGTC TCC TCC CAG CA-3′) [40]. PCR was carried out in a Thermal Cycler Techne® TC-512, in a final volume reaction of 15 μL containing 50 ng DNA, 0.2 mM dNTPs, 1.5 mM MgCl2, 1X PCR buffer +(NH4)2SO4 (Fermentas, MD, USA), 0.4 μM primers, and 1 U Hot Start Taq DNA Polymerase (Fermentas, MD, USA). Reaction mixtures were preheated (94 °C, 5 min) and were followed by PCR with 40 cycles of amplification (94 °C, 60 s; 62.8 °C, 60 s; and 72 °C, 60 s), followed by a final extension at 72 °C during 7 min. Finally, the PCR products (201 bp) were analyzed on 2 % agarose gel containing 10 μg⁄mL ethidium bromide and visualized by UV transillumination. Positive controls of T and C alleles and negative controls (without DNA and feminine DNA) were included in all analysis.
Testicular biopsy
A small piece of testicular tissue was fixed in Bouin’s solution during 6 h for histopathological evaluation. Testicular histology assessment included a qualitative and quantitative analysis of germinal epithelium in 20–25 tubules, the modified Johnsen score (JS) was calculated [20, 21]. According to this score, the tissues were classified in Sertoli cell-only syndrome, complete (JS = 2) or incomplete (some foci of spermatogenesis); Maturation arrest (germ cells until spermatogonia or spermatocyte, which may be complete or incomplete); Hypospermatogenesis (proportional and quantitative reduction of the different types of germ cells); Severe atrophy (hyalinization of seminiferous tubules and lack Sertoli and germ cell, JS = 1); Mixed atrophy (mixture of the above mentioned types of tubular histology); and normal spermatogenesis (all the tubules evaluated had complete spermatogenesis or elongated spermatids at least, JS ≥ 8).
Statistical analysis
Statistical calculations were performed using SPSS 11.5 for Windows (SPSS Inc, Chicago, Illinois). The Pearson Chi square and exact Fisher tests were used for testing differences in proportions between groups. For studying possible differences in means or medians, the groups were compared by the ANOVA and Student’s t test or by the Kruskal–Wallis and Mann–Whitney test, respectively. The Bonferroni test was performed to adjust the P-value for multiple comparisons, specifically when the T and C alleles distributions were compared among controls and 2 groups of patients with spermatogenic failure (with and without Y chromosome microdeletions). P values less than 0.05 (two sided) were considered statistically significant.
Results
The analysis of testicular histology in 252 infertile patients with indication of testicular biopsy allowed us to identify 186 men with spermatogenic impairment of different histological types, and 66 infertile men with complete spermatogenesis (obstructive azo/oligozoospermia). Because no significant differences were observed in the hormonal parameters between obstructive azoospermic, fertile and normozoospermic controls, they were analyzed as a single group (Table 1). The hormonal characterization and the prevalence of reduced testicular volume and azoospermia among secretory azo/oligozoospermic and control men are also shown in Table 1.
Table 1.
Hormonal levels, testicular volume and seminal features in secretory azo/oligozoospermic and control men
| n | FSHa (mIU/ml) | LHa (mIU/ml) | Testosteronea ng/ml | % of patients with testes <30 ccb | Sperm countc mill/mL | % of men with azoospermia | |
|---|---|---|---|---|---|---|---|
| Secretory azo/oligozoospermic group | 218 | ||||||
| Sertoli Cell Only Syndrome (SCOS) | 102 | 15.2 (5.4–44.5)e,g | 5.1 (1.5–14.5)e,g | 3.3 (1.6–6.1)e | 67e | 0.03 ± 0.16 (0–0.8) | 89e,l |
| Maturation Arrest (MA) | 38 | 7.7 (1.8–25.9)e,g | 3.4 (1.3–13.8)e,g | 3.3 (1.8–7.1) | 46e | 0.11 ± 0.47 (0–5) | 73e |
| Mixed Atrophy (MxA) | 20 | 14.4 (2.7–38.3)e,h | 4.8 (1.7–21.1)e,g | 3.0 (0.9–5.6) | 78e | 0.34 ± 1.17 (0–5) | 90e |
| Hypospermatogenesis (HS) | 16 | 5.2 (1.4–17.7)f,g | 4.2 (1.5–7.3)e,g | 3.9 (1.9–6.3) | 33h,i | 0.54 ± 1.31 (0–5) | 63 |
| Severe Atrophy (SA) | 10 | 30.4 (7.9–67)e | 13.4 (4.0–21.8)e | 2.8 (0.8–4.7)f | 89e | 0 | 100e |
| Without diagnosys by testicular biopsyd (WB) | 32 | 11.7 (3.4–53.3)e,g | 4.6 (1.8–13.6)e,g | 4.1 (2.0–6.7)g | 93e,j,k | 0.83 ± 1.36 (0–5.1) | 44m |
| Countrol groupn | 182 | 3.0 (2.0–4.3) | 2.4 (1.6–3.8) | 3.6 (2.9–4.5) | 7 | 55.8 ± 60.5 (0–218.6) | 32 |
| Total | 400 | ||||||
aMedian (2.5–97.5 percentiles)
bCombined testicular volume
cMillions of spermatozoa/mL, mean ± DS (range)
dElevated FSH and/or combined testicular volume <30 cc
eP < 0.01 vs. control group; fP < 0.05 vs. control group; gP < 0.01 vs. SA; hP < 0.05 vs. SA
iP < 0.05 vs. MxA; jP < 0.05 vs. SCOS; kP < 0.01 vs. DM; lP < 0.01 vs. SCOS; mP < 0.01 vs. MxA, SA
nAzo/oligozoospermic obstructive (n = 66), fertile (n = 17) and normozoospermic men (n = 99)
Hormonal normal ranges: FSH (1.0–7.0 mUI/ml), LH (1.0–8.0 mUI/ml), Testosterone (2.0–8.0 ngml)
After screening for Y chromosome microdeletions in 284 azo/oligozoospermic infertile men, we detected 23 non-obstructive azo/oligospermic patients with Y chromosome microdeletions in one (AZFa, AZFb or AZFc) or two AZF regions (AZFb+c). In addition, the analysis of AZFc-partial deletions performed in all subjects detected 14 subjects (14/400) with partial-AZFc deletions, which were “gr/gr” (6 secretory azo/oligozoospermic, 3 obstructive controls and 1 normozoospermic), “b2/b3” (3 secretory azo/oligozoospermic) or “b1/b3” (1 secretory azo/oligozoospermic). AZFc-partial deletions were mainly associated with the absence of DAZ1/DAZ2 (64 %, 9/14), and only two subjects with “gr/gr” (1 secretory azoospermic and 1 obstructive control) and the three patients with “b2/b3” deletions had absence of DAZ3/DAZ4 copies of DAZ gene. After we compared AZFc-partial deletions (total, different subtypes and “gr/gr” with reduction of DAZ1-DAZ2 or DAZ3-DAZ4), no significant differences in the prevalence of AZFc-partial deletions were observed between secretory azo/oligozoospermic patients and controls.
The distribution of T allele (Q1a3a haplogroup) on DYS199 locus initially was studied in secretory azo/oligozospermic men and controls without AZFc-partial deletions (Table 2). When we compared the secretory azo/oligospermic men and controls (total or each subgroup), we observed a similar proportion of Q1a3a haplogroup. However, we observed a higher proportion of the Q1a3a haplogroup in the group of men Y-microdeleted compared to patients with spermatogenic failure without microdeletions of Y chromosome (P = 0.03 by Bonferroni test) or controls (P = 0.017 by Bonferroni test).
Table 2.
Y chromosome Q1a3a haplogroup in cases and controls without AZFc partial deletions
| n | Q1a3a haplogroup* number of patients (%) | |
|---|---|---|
| Secretory Azo/oligozoospermic group | 208 | 16 (7.8) |
| Non Y-microdeleted | 185 | 11 (5.9) |
| Y-microdeleted | 23 | 6 (26)b,c |
| AZFa | 2 | 1 (50) |
| AZFb | 2 | 2 (100)d,e,f |
| AZFc | 13 | 1 (7.7) |
| AZFb+c | 6 | 2 (33.3) |
| Control group | 178 | 16 (9.0) |
| Azo/oligozoospermic obstructive men | 63 | 6 (9.5) |
| Fertile men | 17 | 2 (11.8) |
| Normozoospermic men | 98 | 8 (8.2) |
aDetermined by T allele on DYS1999 locus
bP < 0.01 compared to cases non Y-microdeleted, Bonferroni test
cP < 0.05 compared to total, Bonferroni test
dP < 0.01 compared to cases non Y-microdeleted, total controls or control subgroups, Bonferroni test
eP < 0.01 compared to AZFc Y-microdeleted cases subgroup, Bonferroni test
fP < 0.05 compared to AZFb+c Y-microdeleted cases subgroup, Bonferroni test
Among the Y-microdeleted men, the complete AZFb deletions had an increased prevalence of Q1a3a haplogroup compared to those with AZFc or AZFb+c deletions (P = 0.00009 and P = 0.027 respectively, Bonferroni test). In addition, Y-microdeleted patients with AZFb deletions had an increased prevalence of the Q1a3a haplogroup compared to total controls, control subgroups or secretory azo/oligospermic non Y-microdeleted men (P < 0.01, by Bonferroni test). Among the 14 subjects with partial-AZFc deletions 1 obstructive azoospermic control men with a “gr/gr” subdeletion without DAZ1/DAZ2 had the T allele (Q1a3a haplogroup) and all the remaining had the C allele.
While all subjects in our study were Chilean, and Chilean descendants, we analyzed possible ethnic differences by analyzing their place of residence in Chile, either in different geographical regions or microgeographic Districts within the Metropolitan region (Region of the capital city). This analysis showed that 78 % (300/386) of patients lived in the Metropolitan region (255/386; 66 % living in Santiago), and the remaining lived outside of the Metropolitan region, in the north (8 %) or south (14 %) of the country. When we compared the two groups of secretory azo/oligozoospermic patients, with and without Y chromosome microdeletion, and obstructive controls, no significant differences were observed in the proportion of subjects from different regions of Chile or from different Districts within Santiago or the Metropolitan region. Similar Districts in Santiago or the Metropolitan region were observed between subgroups of controls. However, normozoospermic and fertile controls were predominantly from the Metropolitan region or from Santiago (99 % and 96 %, respectively).
Discussion
In this study we show for the first time that Chilean patients with microdeletions of the Y chromosome have an increased proportion of Y chromosomes belonging to the Q1a3a lineage.
The T allele of DYS199 (M3) locus defines the Q1a3a lineage which represents the prevalent Y chromosome lineage for the native population and their descendants in Chile and the rest of South America [2, 5, 18, 22, 23, 40, 46]. Several studies based on DNA mitochondrial and autosomal microsatellite markers are consistent with a founder effect occurring within the North American subcontinent, before the peopling of Central and South America [30, 42]. Previous studies in the Chilean population of Santiago have shown a prevalence of 84 % of indigenous mitochondrial haplogroups [35], around 40 % of autosomal markers, depending on socioeconomic status, and a smaller proportion (<20 %) of molecular markers of the Y chromosome [8], indicating that the Chilean population arose mainly of the union of indigenous women with European males.
Similar to other studies, we observed that microdeletions that involve AZFa and AZFb had a lower frequency compared to those of AZFc or AZFb+c regions [13, 19, 36, 41]. In addition, similar to a previous study in Chilean patients [29], we did not find a higher proportion of AZFc-partial deletions (“gr/gr”, “b1/b3” and/or “b2/b3”) in secretory azo/oligozoospermic patients compared to controls, and therefore these subdeletions do not seem predispose to severe spermatogenic impairment. However, a recent meta-analysis has shown that “gr/gr” subdeletions can be considered a risk factor for spermatogenic failure especially in oligozoospermics men from Europe [39]. Therefore, the lack of association of AZFc-partial deletions with the primary spermatogenic failure observed in our study may be the consequence of the geographic origin and of the severity of the spermatogenic impairment among our patients (78 % of azoospermia and 54 % of patient had SCOS).
In this study we observed that Chilean men who have Y chromosomes of the Q1a3a haplogroup have a higher proportion of microdeletions in this chromosome. Surprisingly, patients with the more frequent AZF region deleted had a lower proportion or an absence of Q1a3a lineage (complete and AZFc-partial deletions, respectively). In contrast, our two patients with AZFb deletions had a Y chromosome of the Q1a3a lineage, which seems to indicate an increased susceptibility of this South American native haplogroup for AZFb deletions, however additional subjects should be studied. In addition, we observed that patients with AZFb+c or AZFa microdeletions had the T allele, but in a lower proportion to those patients with AZFb microdeletions.
The Y chromosome reference of the GenBank data base shows that Y-M3 SNP (also named as DYS199, rs3894 or sY-103) is localized between the AZFa and AZFb regions, on the chromosomal position 19096363 (GRCh37.p5 Assembly). Interestingly, the T allele was observed in our two patients with AZFb and in one- third of patients with AZFb+c deletions, suggesting that subjects with the T allele on DYS199 locus would have a Y chromosome with a particular sequence variation, or a particular structure that predispose to suffer AZFb microdeletions. Since our two patients with AZFb deletions and the Q1a3a haplogroup have the previously reported P5/proximal-P1 and P4/proximal-P1 deletion, the mechanism involved would increase the susceptibility to homologous recombination between arms of palindromes P5 or P4 and the arm P1.2 of the P1 palindrome [11, 27, 34]. Therefore, some sequence variations associated with the T allele on DYS199 locus, likely localized within or between the P5/P4 and P1 palindromes might lead to a higher AZFb deletion susceptibility.
In agreement with the chromosomal position of the DYS199 locus (GRCh37.p5 Assembly) we detected three patients who had failure in their PCR result for Y-M3 SNP (data not shown) and coincidently their AZFb+c deletions were more proximal (XKRY, CDY2, sY1227 and sY1228 not detected) than those patients in this study (XKRY, CDY2 and sY1228 detected).
Since we observed similar residence places between our infertile patients (Y-microdeleted, non Y-microdeleted and obstructive controls), the statistically significant increased prevalence of the Q1a3a haplogroup in the total Y-microdeleted group or in the AZFb subgroup cannot be attributed to differences in their geographic localization. Therefore, our results suggest a greater susceptibility of the Y chromosomes of Q1a3a South Amerindian lineage to have AZFb or non AZFc deletions. On the other hand, in our Chilean population the AZFc-partial deletions (“gr/gr”, “b1/b3” and/or “b2/b3”) do not seem to predispose to severe spermatogenic impairment.
Acknowledgments
We would like to thank all the men who generously accepted to participate in the study.
This work was supported by the Grant no. 1060081, National Fund for the Scientific and Technological Development (FONDECYT) of Chile.
Footnotes
Capsule
The Q1a3a South Amerindian lineage, defined by a C>T mutation in the DYS199 locus on the Y chromosome, seems to increase the susceptibility to AZF microdeletions in a Chilean population of infertile men.
References
- 1.Arredi B, Ferlin A, Speltra E, Bedin C, Zuccarello D, Ganz F, et al. Y-chromosome haplogroups and susceptibility to azoospermia factor c microdeletion in an Italian population. J Med Genet. 2007;44(3):205–208. doi: 10.1136/jmg.2006.046433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailliet G, Ramallo V, Muzzio M, Garcia A, Santos MR, Alfaro EL, et al. Brief communication: restricted geographic distribution for Y-Q* paragroup in South America. Am J Phys Anthropol. 2009;140(3):578–582. doi: 10.1002/ajpa.21133. [DOI] [PubMed] [Google Scholar]
- 3.Bhasin S. Approach to the infertile man. J Clin Endocrinol Metab. 2007;92(6):1995–2004. doi: 10.1210/jc.2007-0634. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi NO, Bailliet G, Bravi CM, Carnese RF, Rothhammer F, Martinez-Marignac VL, et al. Origin of Amerindian Y-chromosomes as inferred by the analysis of six polymorphic markers. Am J Phys Anthropol. 1997;102(1):79–89. doi: 10.1002/(SICI)1096-8644(199701)102:1<79::AID-AJPA7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Blanco-Verea A, Jaime JC, Brion M, Carracedo A. Y-chromosome lineages in native South American population. Forensic Sci Int Genet. 2010;4(3):187–193. doi: 10.1016/j.fsigen.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Carrell DT. Elucidating the genetics of male infertility: understanding transcriptional and translational regulatory networks involved in spermatogenesis. Int J Androl. 2008;31(5):455–456. doi: 10.1111/j.1365-2605.2008.00913.x. [DOI] [PubMed] [Google Scholar]
- 7.Castro A, Zambrano N, Kaune H, Madariaga M, Lopez P, Mericq V. YqTER deletion causes arrest of spermatogenesis in early puberty. J Pediatr Endocrinol Metab. 2004;17(12):1675–1678. doi: 10.1515/JPEM.2004.17.12.1675. [DOI] [PubMed] [Google Scholar]
- 8.Cifuentes L, Morales R, Sepulveda D, Jorquera H, Acuna M. DYS19 and DYS199 loci in a Chilean population of mixed ancestry. Am J Phys Anthropol. 2004;125(1):85–89. doi: 10.1002/ajpa.10380. [DOI] [PubMed] [Google Scholar]
- 9.Cifuentes L, Valenzuela CY, Cruz-Coke R, Armanet L, Lyng C, Harb Z. Genetic characterization of the hospital population of Santiago, Chile. Rev Med Chil. 1988;116(1):28–33. [PubMed] [Google Scholar]
- 10.Consortium Y-C. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12(2):339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa P, Goncalves R, Ferras C, Fernandes S, Fernandes AT, Sousa M, et al. Identification of new breakpoints in AZFb and AZFc. Mol Hum Reprod. 2008;14(4):251–258. doi: 10.1093/molehr/gan014. [DOI] [PubMed] [Google Scholar]
- 12.Ferlin A, Arredi B, Foresta C. Genetic causes of male infertility. Reprod Toxicol. 2006;22(2):133–141. doi: 10.1016/j.reprotox.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, et al. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92(3):762–770. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- 14.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14(6):734–745. doi: 10.1016/S1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes AT, Fernandes S, Goncalves R, Sa R, Costa P, Rosa A, et al. DAZ gene copies: evidence of Y chromosome evolution. Mol Hum Reprod. 2006;12(8):519–523. doi: 10.1093/molehr/gal051. [DOI] [PubMed] [Google Scholar]
- 16.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22(2):226–239. doi: 10.1210/er.22.2.226. [DOI] [PubMed] [Google Scholar]
- 17.Gianotten J, Lombardi MP, Zwinderman AH, Lilford RJ, van der Veen F. Idiopathic impaired spermatogenesis: genetic epidemiology is unlikely to provide a short-cut to better understanding. Hum Reprod Update. 2004;10(6):533–539. doi: 10.1093/humupd/dmh045. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Neira A, Gusmao L, Brion M, Lareu MV, Amorim A, Carracedo A. Distribution of Y-chromosome STR defined haplotypes in Iberia. Forensic Sci Int. 2000;110(2):117–126. doi: 10.1016/S0379-0738(00)00156-0. [DOI] [PubMed] [Google Scholar]
- 19.Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18(8):1660–1665. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- 20.Jezek D, Knuth UA, Schulze W. Successful testicular sperm extraction (TESE) in spite of high serum follicle stimulating hormone and azoospermia: correlation between testicular morphology, TESE results, semen analysis and serum hormone values in 103 infertile men. Hum Reprod. 1998;13(5):1230–1234. doi: 10.1093/humrep/13.5.1230. [DOI] [PubMed] [Google Scholar]
- 21.Johnsen SG. Testicular biopsy score count–a method for registration of spermatogenesis in human testes: normal values and results in 335 hypogonadal males. Hormones. 1970;1(1):2–25. doi: 10.1159/000178170. [DOI] [PubMed] [Google Scholar]
- 22.Jota MS, Lacerda DR, Sandoval JR, Vieira PP, Santos-Lopes SS, Bisso-Machado R, et al. A new subhaplogroup of native American Y-Chromosomes from the Andes. Am J Phys Anthropol. 2011;146(4):553–559. doi: 10.1002/ajpa.21519. [DOI] [PubMed] [Google Scholar]
- 23.Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18(5):830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krausz C, Bussani-Mastellone C, Granchi S, McElreavey K, Scarselli G, Forti G. Screening for microdeletions of Y chromosome genes in patients undergoing intracytoplasmic sperm injection. Hum Reprod. 1999;14(7):1717–1721. doi: 10.1093/humrep/14.7.1717. [DOI] [PubMed] [Google Scholar]
- 25.Krausz C, Giachini C. Genetic risk factors in male infertility. Arch Androl. 2007;53(3):125–133. doi: 10.1080/01485010701271786. [DOI] [PubMed] [Google Scholar]
- 26.Kruger TF, Ackerman SB, Simmons KF, Swanson RJ, Brugo SS, Acosta AA. A quick, reliable staining technique for human sperm morphology. Arch Androl. 1987;18(3):275–277. doi: 10.3109/01485018708988493. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29(3):279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 28.Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138(5):855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lardone MC, Parodi DA, Ebensperger M, Penaloza P, Cornejo V, Valdevenito R, et al. AZFc partial deletions in Chilean men with severe spermatogenic failure. Fertil Steril. 2007;88(5):1318–1326. doi: 10.1016/j.fertnstert.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 30.Lewis CM., Jr Hierarchical modeling of genome-wide Short Tandem Repeat (STR) markers infers native American prehistory. Am J Phys Anthropol. 2010;141(2):281–289. doi: 10.1002/ajpa.21143. [DOI] [PubMed] [Google Scholar]
- 31.Ma K, Mallidis C, Bhasin S. The role of Y chromosome deletions in male infertility. Eur J Endocrinol. 2000;142(5):418–430. doi: 10.1530/eje.0.1420418. [DOI] [PubMed] [Google Scholar]
- 32.Navarro-Costa P, Plancha CE, Goncalves J. Genetic dissection of the AZF regions of the human Y chromosome: thriller or filler for male (in)fertility? J Biomed Biotechnol. 2010;2010:936569. doi: 10.1155/2010/936569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online. 2008;16(4):504–513. doi: 10.1016/S1472-6483(10)60457-9. [DOI] [PubMed] [Google Scholar]
- 34.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71(4):906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocco P, Morales C, Moraga M, Miquel JF, Nervi F, Llop E, et al. Genetic composition of the Chilean population. Analysis of mitochondrial DNA polymorphism. Rev Med Chil. 2002;130(2):125–131. [PubMed] [Google Scholar]
- 36.Sadeghi-Nejad H, Farrokhi F. Genetics of azoospermia: current knowledge, clinical implications, and future directions. Part II: Y chromosome microdeletions. Urol J. 2007;4(4):192–206. [PubMed] [Google Scholar]
- 37.Silber SJ. The Y chromosome in the era of intracytoplasmic sperm injection: a personal review. Fertil Steril. 2011;8:2439–48. doi: 10.1016/j.fertnstert.2011.05.070. [DOI] [PubMed] [Google Scholar]
- 38.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 39.Stouffs K, Lissens W, Tournaye H, Haentjens P. What about gr/gr deletions and male infertility? systematic review and meta-analysis. Hum Reprod Update. 2011;17(2):197–209. doi: 10.1093/humupd/dmq046. [DOI] [PubMed] [Google Scholar]
- 40.Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci U S A. 1996;93(1):196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogt P. Human chromosome deletions in Yq11, AZF candidate genes and male infertility: history and update. Mol Hum Reprod. 1998;4(8):739–744. doi: 10.1093/molehr/4.8.739. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3(11):e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laboratory manual for the examination of human semen and sperm-cervical mucus interaction. 4. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 44.Yang Y, Ma M, Li L, Zhang W, Chen P, Ma Y, et al. Y chromosome haplogroups may confer susceptibility to partial AZFc deletions and deletion effect on spermatogenesis impairment. Hum Reprod. 2008;23(9):2167–2172. doi: 10.1093/humrep/den229. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Ma M, Li L, Zhang W, Xiao C, Li S, et al. Evidence for the association of Y-chromosome haplogroups with susceptibility to spermatogenic failure in a Chinese Han population. J Med Genet. 2008;45(4):210–215. doi: 10.1136/jmg.2007.054478. [DOI] [PubMed] [Google Scholar]
- 46.Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol. 2004;21(1):164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F, Lu C, Li Z, Xie P, Xia Y, Zhu X, et al. Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet. 2007;44(7):437–444. doi: 10.1136/jmg.2007.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]