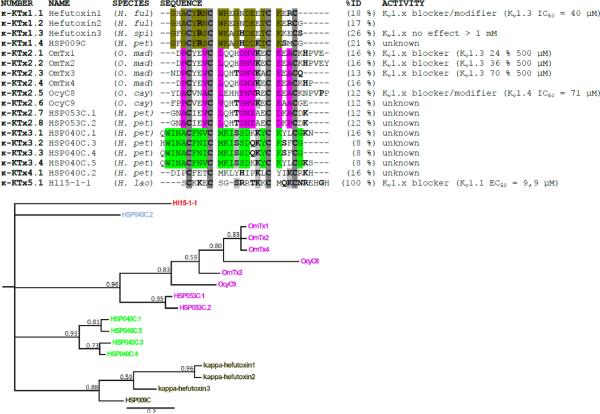
Fig. 3.
Multiple sequence alignment of Hl15-1-1 with the other existing κ-KTx family members. %ID is the identity score of the peptide compared to Hl15-1-1. The alignment is based on the cysteine residues (bold and dark gray background). The bold residues in a light gray background are amino acids identical to Hl15-1-1 and other κ-KTx toxins. Bold residues represent those amino acids having a side chain with a similar chemical nature. For each suggested κ-KTx subfamily, those residues having the same chemical properties are colored. The multiple sequence alignment has been used for constructing a tree topology by Bayesian inference. The consensus tree shown supports the suggested division of the κ-KTx family. The posterior probability indicating the clade credibility is shown at each node. The scale bar gives the expected changes per amino acid site. Species names are H. ful (Heterometrus fulvipus), H. spi (Heterometrus spinnifer), O. mad (Opisthacanthus madagascariensis), O. cay (Opisthacanthus cayaporum), H. pet (Heterometrus petersii) and H. lao (Heterometrus laoticus).