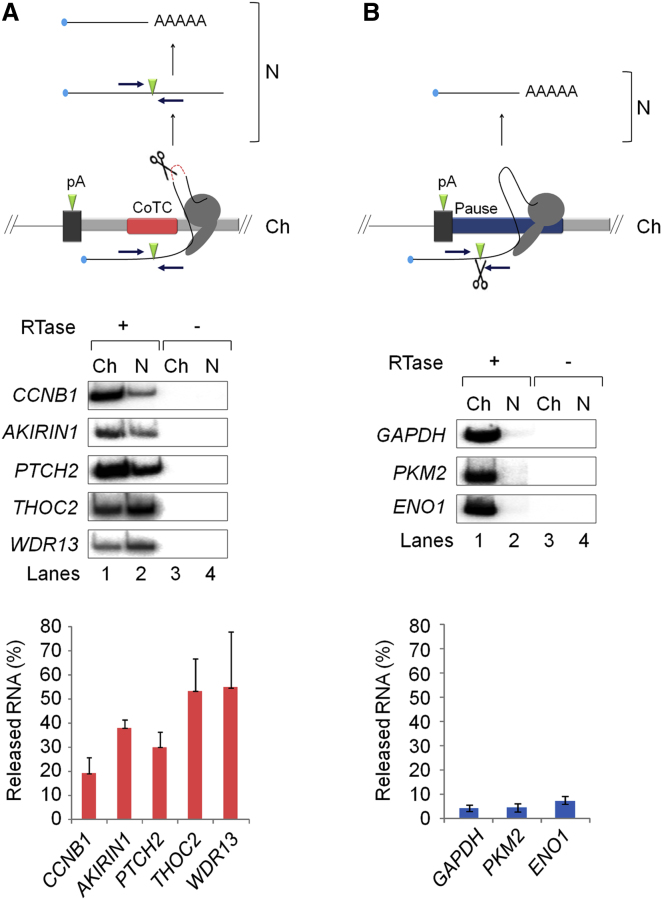
Figure 2.
Nuclear Distribution of Pre-mRNA from CstF-64 CLIP Positive and Negative Gene Loci
CLIP positive (A) and negative (B) gene names are shown to the left of the corresponding data panels. RT-PCR analysis was performed on pre-mRNA from chromatin (Ch, lanes 1 and 3) and nucleoplasm (N, lanes 2 and 4) fractions. cDNA synthesis was primed using random primers, in reactions with (+RTase, lanes 1 and 2) or without (−RTase, lanes 3 and 4) the addition of reverse transcriptase to control for the presence of contaminating DNA. PCR amplification (23 cycles) of the resulting cDNA was conducted using gene-specific primers (indicated by blue arrows in the diagrams above the data panels) spanning annotated poly(A) sites (UCSC Genome Browser; http://genome.ucsc.edu/). (The number of PCR cycles was determined to be within the linear range [data not shown] and therefore accurately reflects RNA abundance). Radiolabeled RT-PCR products from chromatin and nucleoplasm fractions (lanes 1 and 2) were quantitated by PhosphoImage analysis and the proportion of released nucleoplasmic pre-mRNA (% total) calculated and displayed in the graphs below the data panels. The lack of PCR products in lanes 3 and 4 (−RTase) confirms the absence of contaminating DNA in all samples. The diagrams above the data panels illustrating CoTC-type (A) and pause-type (B) termination mechanisms are labeled as Figure 1A, except for the pause element (blue bar) and scissors (indicating cleavage by the 3′ end processing complex) in (B). Error bars represent the results of three experimental repeats.