Figure S1.
Control Experiment to Test RT-PCR Efficiency on PCTE Transcripts, Related to Figure 3
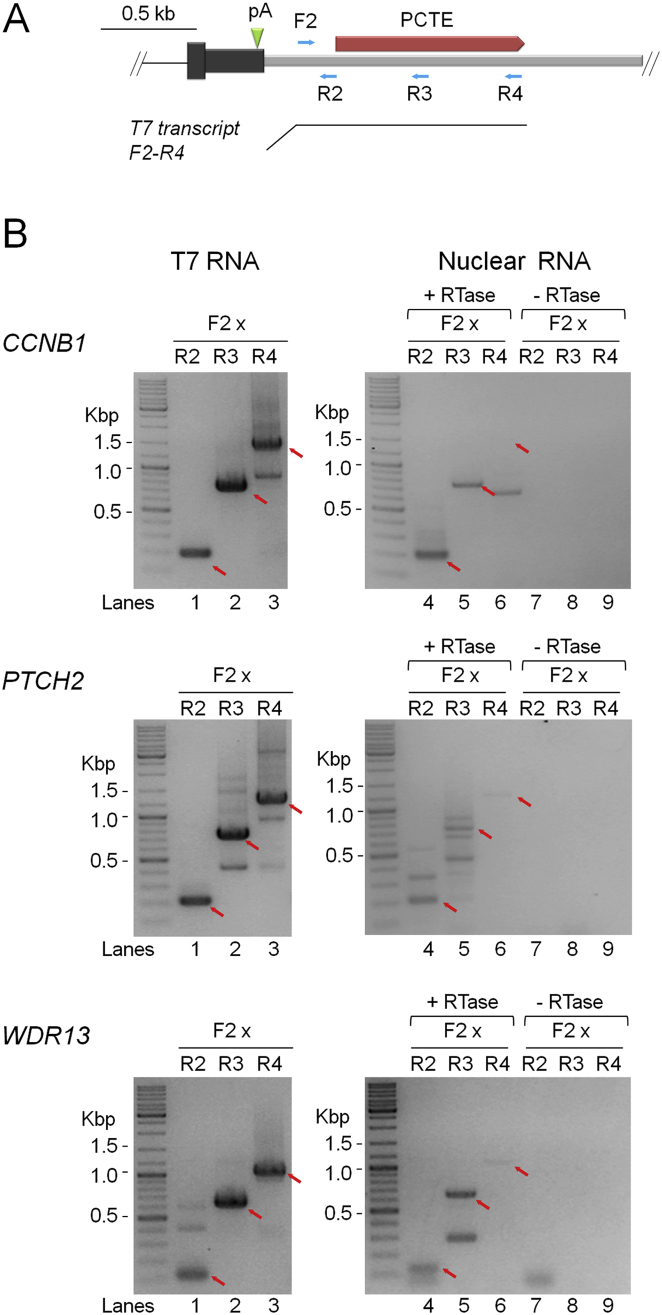
(A) Diagram showing candidate gene PCTEs (red box), control T7 transcripts (black line) and PCR primers (blue arrows). The gene specific PCR primer sets used are as in Figure 3.
(B) Data from RT-PCR analysis of CCNB1, PTCH2 and WDR13 PCTEs. Full length T7 PCTE transcripts and PCTE transcripts from HeLa cell nuclei were reverse transcribed with random primers. The resulting cDNAs were PCR amplified (26 PCR cycles for T7 RNA templated cDNA and 32 cycles for HeLa nuclear RNA templated cDNA) using indicated gene specific primer pairs. Lanes 1-3, RT-PCR products from T7 PCTE transcripts. The presence of the prominent, expected, PCR products (indicated by red arrows) in each lane indicates that the control T7 PCTE transcripts are efficiently reverse transcribed and PCR amplified, irrespective of their length. Lanes 4-6, RT-PCR products from HeLa nuclear PCTE transcripts. Prominent bands, corresponding to the 5′ end of PCTE transcripts, are detected in lane 4 for all samples, however RT-PCR products representing longer PCTE transcripts, in lanes 5 and 6, are of lower abundance and in some cases entirely absent (e.g., CCNB1, lane 6). Comparison of HeLa nuclear PCTE transcript RT-PCR products (in lanes 4-6) with RT-PCR products from control T7 transcripts (lanes 1-3) shows that detection of longer endogenous nuclear transcripts is significantly lower than for the corresponding T7 control transcripts. This result indicates that the longer PCTE transcripts are less abundant in HeLa nuclei, due to transcript termination or transcript cleavage, rather than template length dependent decrease in RT-PCR efficiency. The absence of PCR products in control lanes 7-9 (-RTase) confirms the absence of contaminating DNA in these experiments.