Abstract
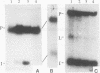
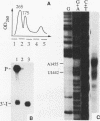
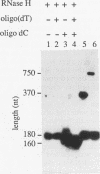
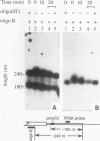
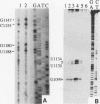
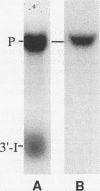
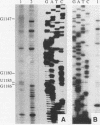
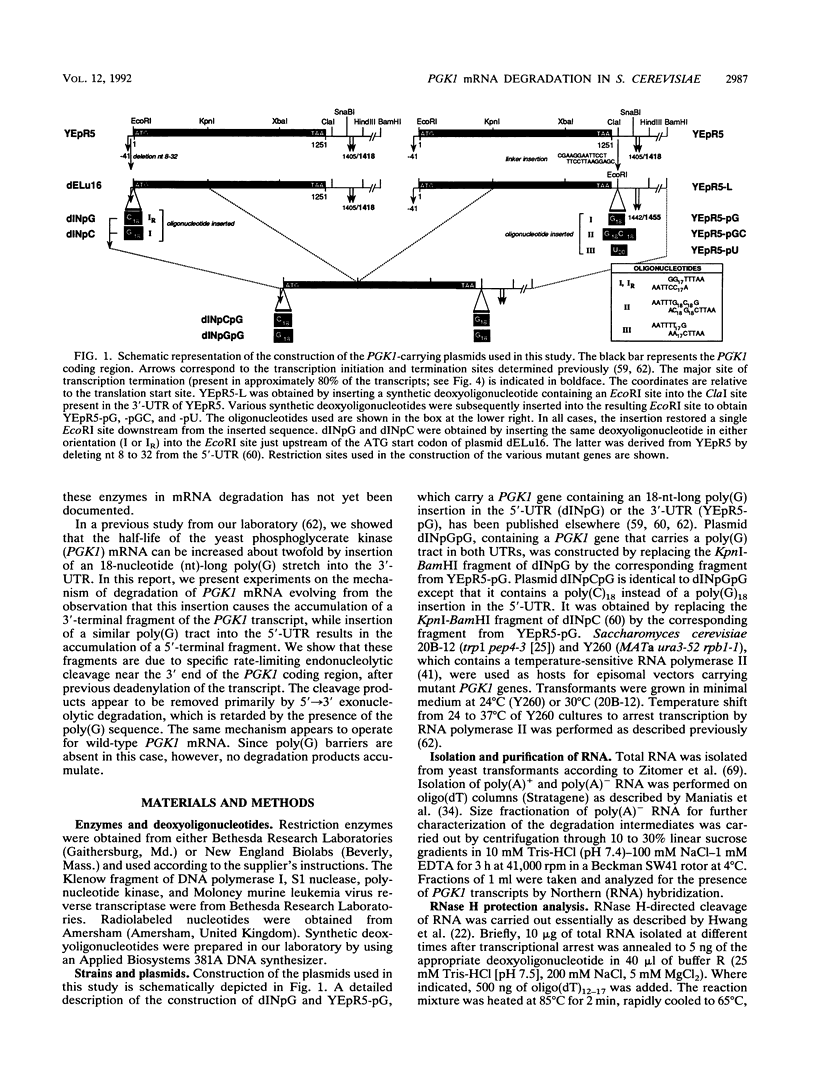
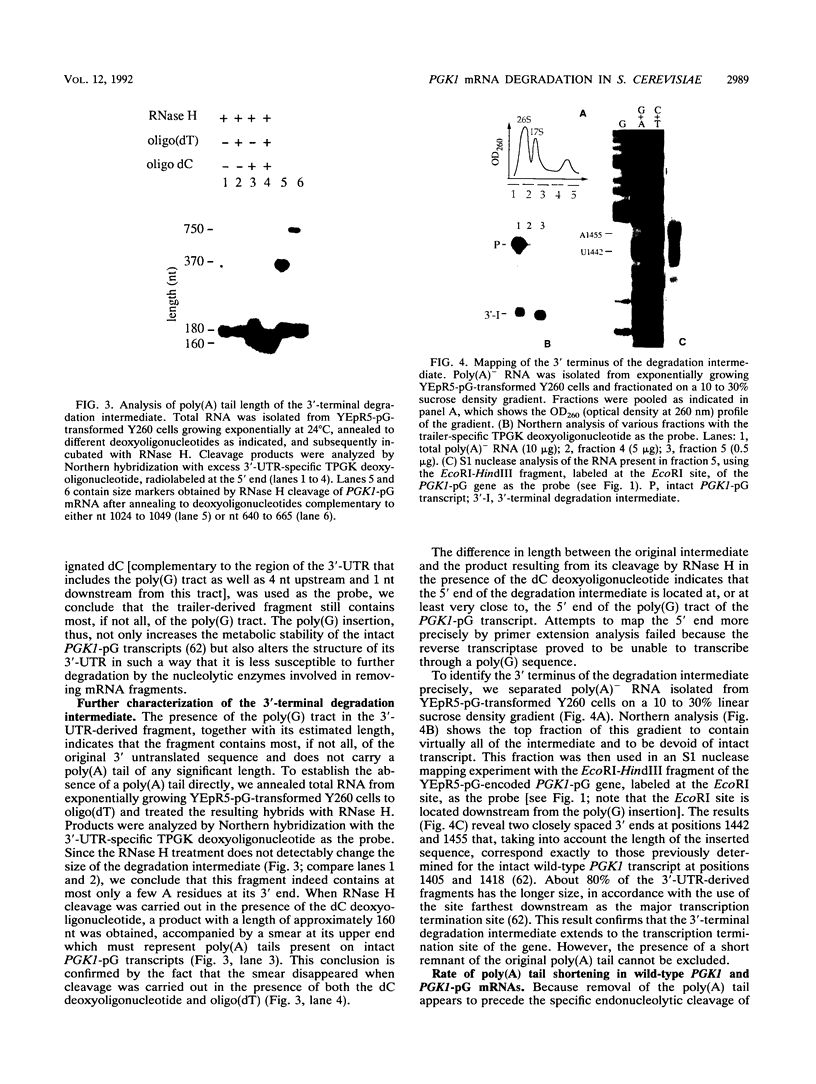
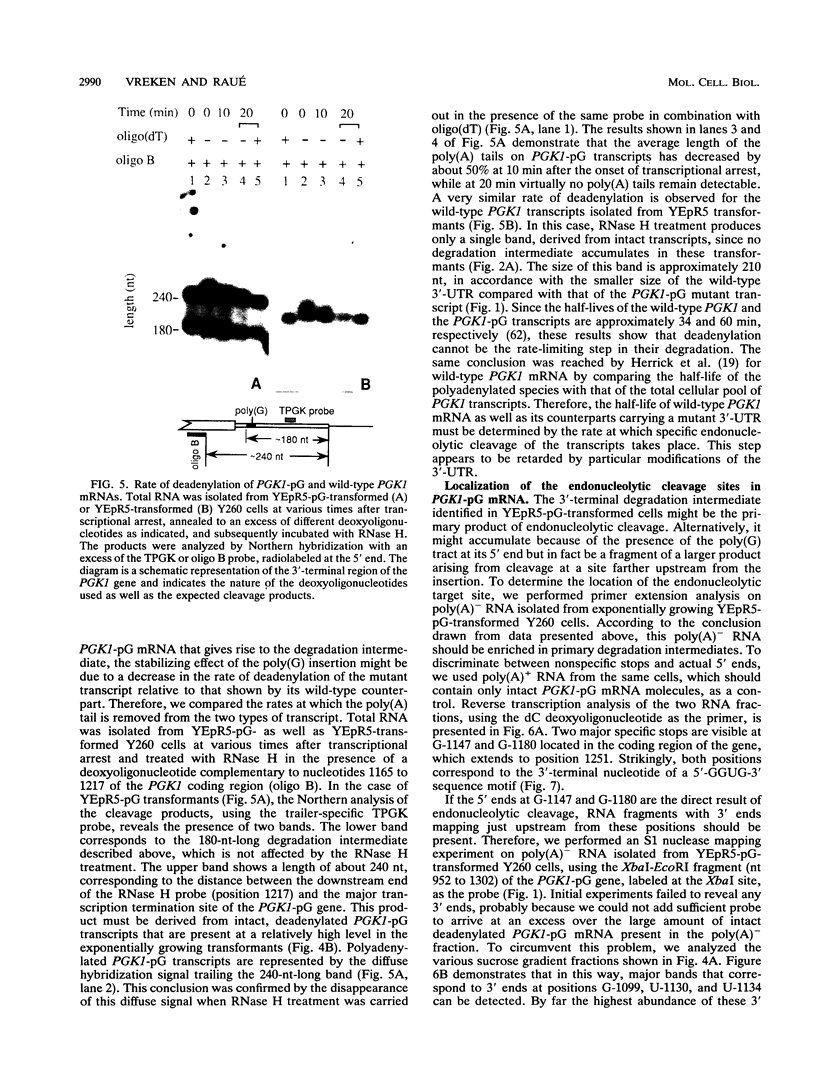
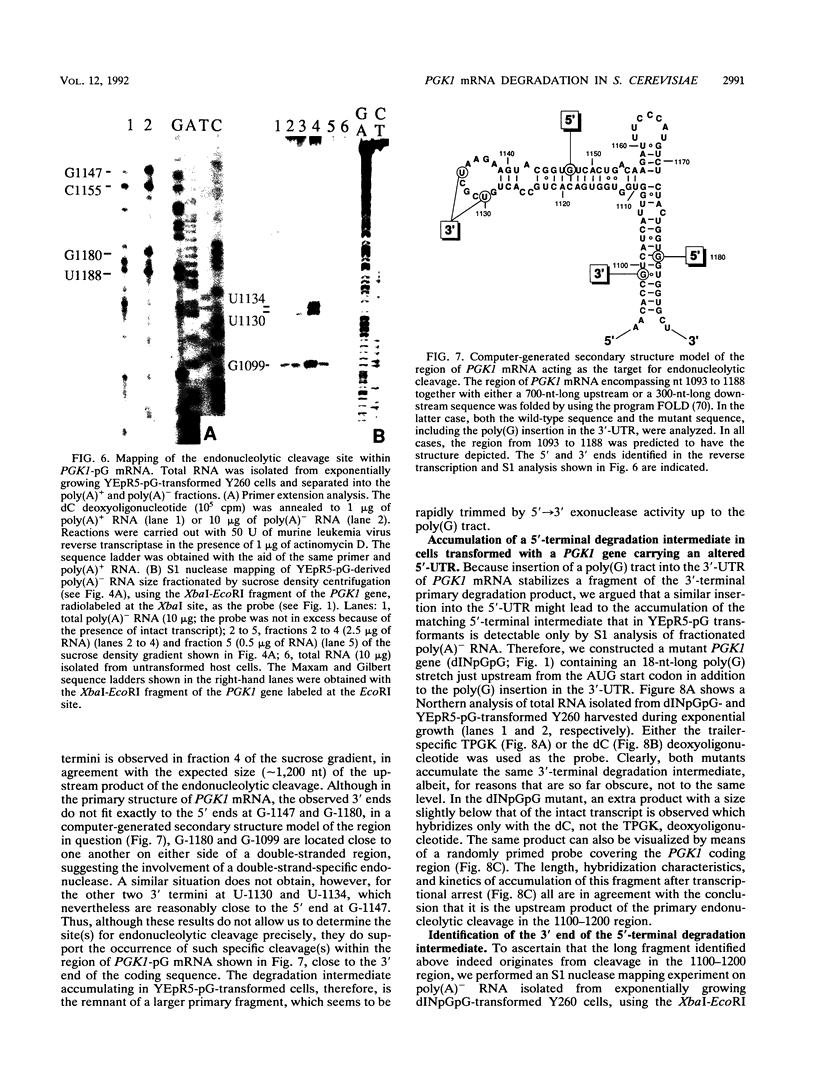
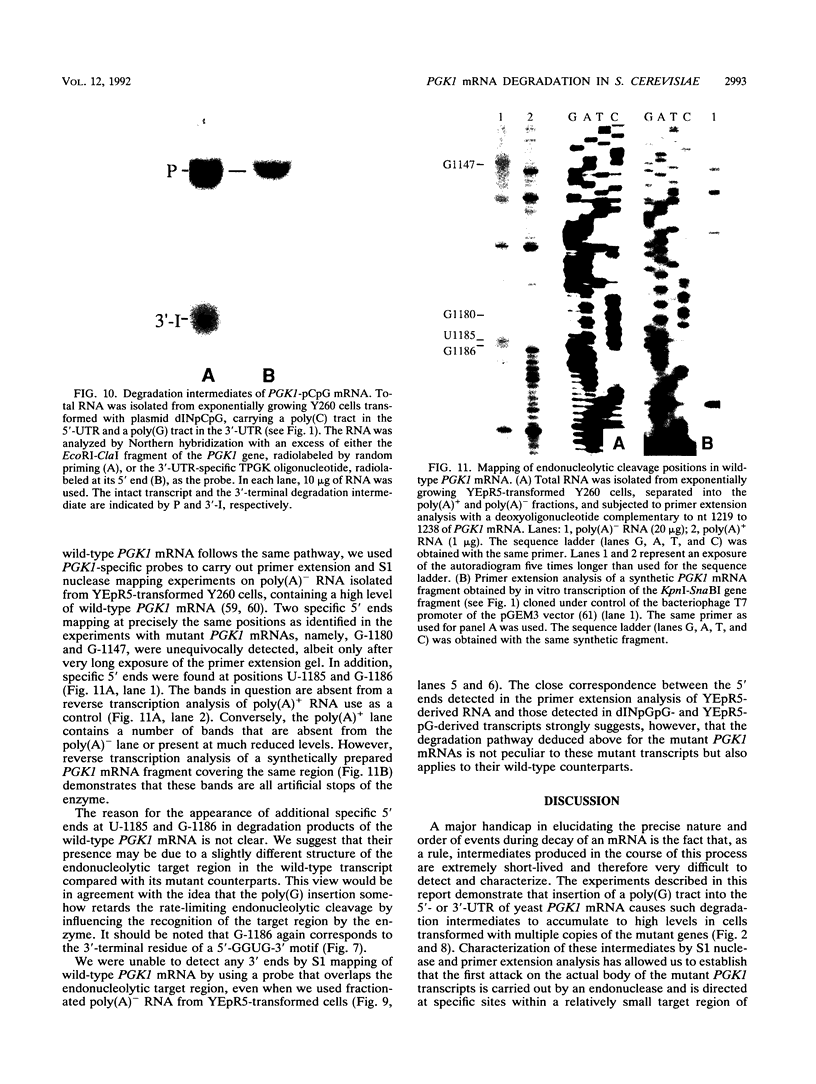
Insertion of an 18-nucleotide-long poly(G) tract into the 3'-terminal untranslated region of yeast phosphoglycerate kinase (PGK1) mRNA increases its chemical half-life by about a factor of 2 (P. Vreken, R. Van der Veen, V. C. H. F. de Regt, A. L. de Maat, R. J. Planta, and H. A. Raué, Biochimie 73:729-737, 1991). In this report, we show that this insertion also causes the accumulation of a degradation intermediate extending from the poly(G) sequence down to the transcription termination site. Reverse transcription and S1 nuclease mapping experiments demonstrated that this intermediate is the product of shorter-lived primary fragments resulting from endonucleolytic cleavage immediately downstream from the U residue of either of two 5'-GGUG-3' sequences present between positions 1100 and 1200 close to the 3' terminus (position 1251) of the coding sequence. Similar endonucleolytic cleavages appear to initiate degradation of wild-type PGK1 mRNA. Insertion of a poly(G) tract just upstream from the AUG start codon resulted in the accumulation of a 5'-terminal degradation intermediate extending from the insertion to the 1100-1200 region. RNase H degradation in the presence of oligo(dT) demonstrated that the wild-type and mutant PGK1 mRNAs are deadenylated prior to endonucleolytic cleavage and that the half-life of the poly(A) tail is three- to sixfold lower than that of the remainder of the mRNA. Thus, the endonucleolytic cleavage constitutes the rate-limiting step in degradation of both wild-type and mutant PGK1 transcripts, and the resulting fragments are degraded by a 5'----3' exonuclease, which appears to be severely retarded by a poly(G) sequence.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atwater J. A., Wisdom R., Verma I. M. Regulated mRNA stability. Annu Rev Genet. 1990;24:519–541. doi: 10.1146/annurev.ge.24.120190.002511. [DOI] [PubMed] [Google Scholar]
- Barker G. F., Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991 May;11(5):2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Bernstein P., Peltz S. W., Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989 Feb;9(2):659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R., Hwang S. P., Ratnasabapathy R., Williams D. L. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5'-AAU-3'/5'-UAA-3' elements in single-stranded loop domains of the 3'-noncoding region. J Biol Chem. 1989 Oct 5;264(28):16910–16918. [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Paris J., Phillippe M., Osborne H. B. Degradation of a developmentally regulated mRNA in Xenopus embryos is controlled by the 3' region and requires the translation of another maternal mRNA. Mol Cell Biol. 1991 Jun;11(6):3115–3124. doi: 10.1128/mcb.11.6.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989 May;9(5):1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J. Messenger RNA stability in yeast. Yeast. 1989 Jul-Aug;5(4):239–257. doi: 10.1002/yea.320050405. [DOI] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J. L., Koeller D. M., Ramin V. C., Klausner R. D., Harford J. B. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 1989 Dec 1;8(12):3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D. A., Sisodia S. S., Cleveland D. W. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalbo D., Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990 Jan;17(1):77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Henderson E., Hardin C. C., Walk S. K., Tinoco I., Jr, Blackburn E. H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987 Dec 24;51(6):899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Hayflick J. S., Chen C. Y., Seeburg P. H., Derynck R. The primary structure of the Saccharomyces cerevisiae gene for 3-phosphoglycerate kinase. Nucleic Acids Res. 1982 Dec 11;10(23):7791–7808. doi: 10.1093/nar/10.23.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A., Kastelein R. A., Vasser M., de Boer H. A. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987 Aug;7(8):2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. P., Eisenberg M., Binder R., Shelness G. S., Williams D. L. Predicted structures of apolipoprotein II mRNA constrained by nuclease and dimethyl sulfate reactivity: stable secondary structures occur predominantly in local domains via intraexonic base pairing. J Biol Chem. 1989 May 15;264(14):8410–8418. [PubMed] [Google Scholar]
- Iwai Y., Bickel M., Pluznik D. H., Cohen R. B. Identification of sequences within the murine granulocyte-macrophage colony-stimulating factor mRNA 3'-untranslated region that mediate mRNA stabilization induced by mitogen treatment of EL-4 thymoma cells. J Biol Chem. 1991 Sep 25;266(27):17959–17965. [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäck H. M., Berg J., Wabl M. Translation affects immunoglobulin mRNA stability. Eur J Immunol. 1989 May;19(5):843–847. doi: 10.1002/eji.1830190510. [DOI] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy I. M., Haddow J. K., Clements J. B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991 Apr;65(4):2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita J. Memory in a neuron. Sci Am. 1989 Jan;260(1):28–32. doi: 10.1038/scientificamerican0189-28. [DOI] [PubMed] [Google Scholar]
- Koeller D. M., Casey J. L., Hentze M. W., Gerhardt E. M., Chan L. N., Klausner R. D., Harford J. B. A cytosolic protein binds to structural elements within the iron regulatory region of the transferrin receptor mRNA. Proc Natl Acad Sci U S A. 1989 May;86(10):3574–3578. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., de Wit C. L., Elfferich P., van der Eb A. J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990 Dec;10(12):6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Nakao Y., Bock R. M. The nucleases of yeast. II. Purification, properties and specificity of an endonuclease from yeast. Biochim Biophys Acta. 1968 Jan 8;151(1):126–136. doi: 10.1016/0005-2744(68)90167-8. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Hagendoorn M. J., Mager W. H., Planta R. J. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984 Sep 11;12(17):6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter J. S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989 Nov 3;246(4930):664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- Mead D. J., Oliver S. G. Purification and properties of a double-stranded ribonuclease from the yeast Saccharomyces cerevisiae. Eur J Biochem. 1983 Dec 15;137(3):501–507. doi: 10.1111/j.1432-1033.1983.tb07854.x. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor B., Bonneaud N., Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol. 1991 Jun;11(6):3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. Tales of poly(A): a review. Gene. 1990 Jul 16;91(2):151–158. doi: 10.1016/0378-1119(90)90082-3. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Neupert B., Kühn L. C. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989 Jul 28;58(2):373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT alpha 1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Ross J. Autogenous regulation of histone mRNA decay by histone proteins in a cell-free system. Mol Cell Biol. 1987 Dec;7(12):4345–4356. doi: 10.1128/mcb.7.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppel K., Vinci J. M., Baglioni C. The AU-rich sequences in the 3' untranslated region mediate the increased turnover of interferon mRNA induced by glucocorticoids. J Exp Med. 1991 Feb 1;173(2):349–355. doi: 10.1084/jem.173.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen D. D., Koch S. R., Granner D. K. 3' noncoding region of phosphoenolpyruvate carboxykinase mRNA contains a glucocorticoid-responsive mRNA-stabilizing element. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7800–7804. doi: 10.1073/pnas.86.20.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Soloway P. D., Shenk T. The adenovirus type 5 i-leader open reading frame functions in cis to reduce the half-life of L1 mRNAs. J Virol. 1990 Feb;64(2):551–558. doi: 10.1128/jvi.64.2.551-558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Maupin M. K. A 5'----3' exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Arch Biochem Biophys. 1987 Feb 1;252(2):339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- Stevens A. Novel specificity of an endoribonuclease of yeast. FEBS Lett. 1986 Sep 15;205(2):210–214. doi: 10.1016/0014-5793(86)80899-7. [DOI] [PubMed] [Google Scholar]
- Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5'-mononucleotides by a 5' leads to 3' mode of hydrolysis. J Biol Chem. 1980 Apr 10;255(7):3080–3085. [PubMed] [Google Scholar]
- Stevens A. Pyrimidine-specific cleavage by an endoribonuclease of Saccharomyces cerevisiae. J Bacteriol. 1985 Oct;164(1):57–62. doi: 10.1128/jb.164.1.57-62.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle M. Y., Hanafusa H. Processing of 9E3 mRNA and regulation of its stability in normal and Rous sarcoma virus-transformed cells. Mol Cell Biol. 1989 Nov;9(11):4738–4745. doi: 10.1128/mcb.9.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C. Regulation of ferritin and transferrin receptor mRNAs. J Biol Chem. 1990 Mar 25;265(9):4771–4774. [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P., van der Veen R., de Regt V. C., de Maat A. L., Planta R. J., Raué H. A. Turnover rate of yeast PGK mRNA can be changed by specific alterations in its trailer structure. Biochimie. 1991 Jun;73(6):729–737. doi: 10.1016/0300-9084(91)90053-4. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Raghuraman M. K., Cech T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989 Dec 1;59(5):871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Xu H. X., Johnson L., Grunstein M. Coding and noncoding sequences at the 3' end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol. 1990 Jun;10(6):2687–2694. doi: 10.1128/mcb.10.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. J., Gay D. A., Pachter J. S., Cleveland D. W. Autoregulated changes in stability of polyribosome-bound beta-tubulin mRNAs are specified by the first 13 translated nucleotides. Mol Cell Biol. 1988 Mar;8(3):1224–1235. doi: 10.1128/mcb.8.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. J., Machlin P. S., Cleveland D. W. Autoregulated instability of beta-tubulin mRNAs by recognition of the nascent amino terminus of beta-tubulin. Nature. 1988 Aug 18;334(6183):580–585. doi: 10.1038/334580a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Montgomery D. L., Nichols D. L., Hall B. D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel J. J., Bergkamp R. J., Planta R. J., Raué H. A. Effect of deletions in the 5'-noncoding region on the translational efficiency of phosphoglycerate kinase mRNA in yeast. Gene. 1989 Jun 30;79(1):83–95. doi: 10.1016/0378-1119(89)90094-2. [DOI] [PubMed] [Google Scholar]
- van den Heuvel J. J., Planta R. J., Raué H. A. Effect of leader primary structure on the translational efficiency of phosphoglycerate kinase mRNA in yeast. Yeast. 1990 Nov-Dec;6(6):473–482. doi: 10.1002/yea.320060604. [DOI] [PubMed] [Google Scholar]