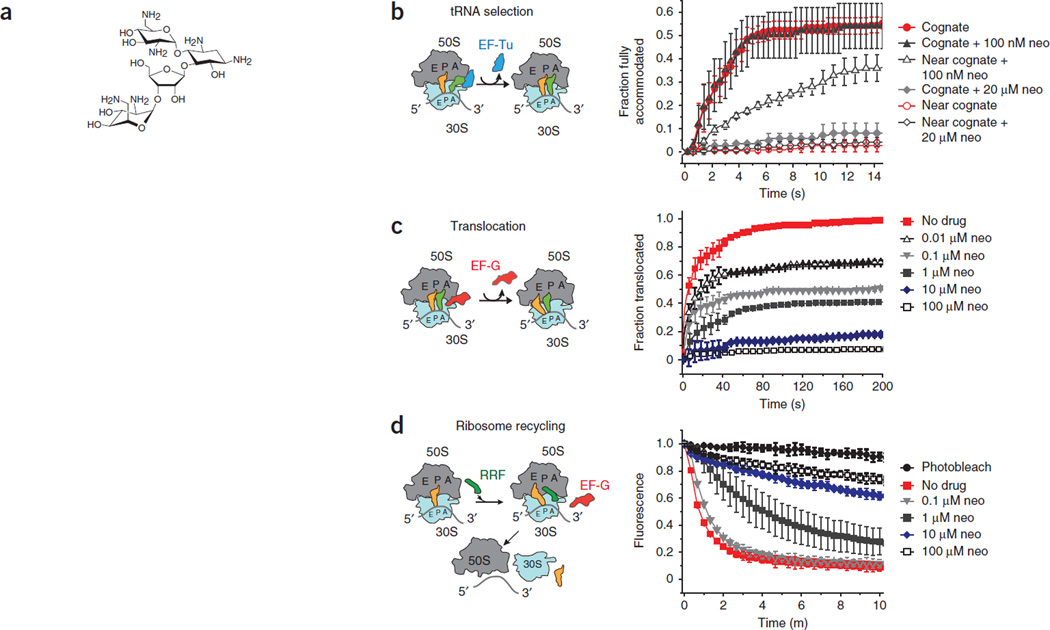
Figure 1.
Neomycin inhibits ribosome functions in vitro. (a) The chemical structure of neomycin. (b) EF-Tu (GTP)-catalyzed accommodation of Phe-tRNAPhe at the A site of wild-type ribosomes programmed with cognate (UUC) and near-cognate (UCU) codons monitored by FRET. The incorporation of cognate tRNA and near-cognate tRNA in the absence of antibiotics is shown. The incorporation of cognate tRNA and near-cognate tRNA in the presence of two concentrations of neomycin (neo) are shown. Experiments were conducted in triplicates, and the mean ± s.d. are plotted. (c) EF-G–catalyzed translocation, monitored by smFRET, normalized to a control group receiving no neomycin (no drug). The fractions of translocated molecules at various concentrations of neomycin are shown. Experiments were conducted in triplicates, and the mean ± s.d. are plotted. (d) Recycling of wild-type 70S ribosome complexes monitored by the disappearance of Cy5-labeled L1 fluorescence in 50S subunits from surface-immobilized wild-type ribosome complexes. Ribosome recycling at various concentrations of neomycin is shown. Under identical conditions, photobleaching was negligible. Experiments were conducted in triplicates, and the mean ± s.d. are plotted.