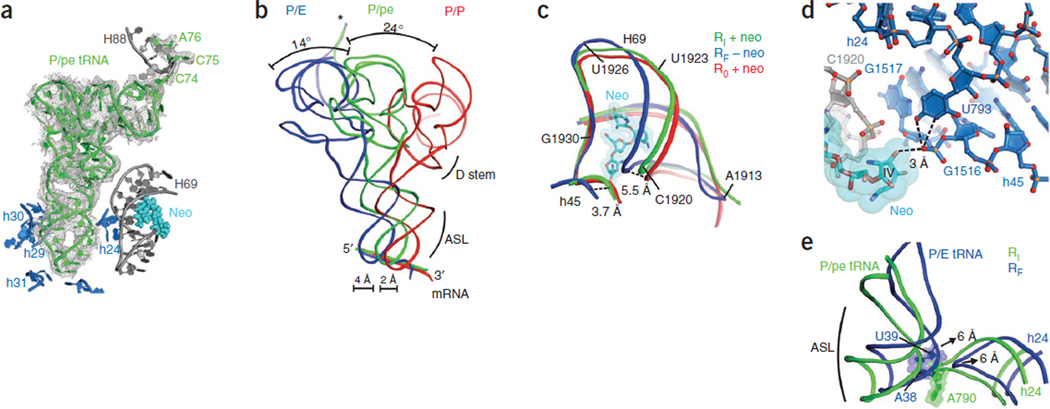
Figure 4.
Position of tRNAPhe in the intermediate-rotated and neomycin-bound ribosome. (a) Electron density map of the Fobs–Fcalc difference for P/pe tRNA in the neomycin-bound, intermediate-rotated ribosome configuration, with its position relative to rRNA elements of the small (blue) and large (gray) subunits and neomycin (light blue spheres) bound to H69 shown. The electron density map is contoured at 2.5 s.d. from the mean. (b) The intermediate position of P-site tRNA (P/pe, green) is between the classical (red) and hybrid (blue) configurations in the extent of both T-stem and anticodon displacement toward the E site. The 3′ CCA end of P/pe tRNA (asterisk) occupies the large-subunit E site. The dihydrouridine (D) stem and the anticodon stem loop (ASL) are indicated. (c) Superposition of the positions of H69 and h45 in unrotated (R0; P/P tRNA), fully rotated (RF; P/E tRNA) and intermediate-rotated (RI; P/pe tRNA) ribosome configurations showing the position of neomycin (neo) in the RI structure. (d) Neomycin (light blue) binding to H69 (gray) stabilizes the small-subunit platform conformation through interactions between neomycin ring IV, the G1517 backbone in h45 (blue) and the U793 residue in h24 (dark blue). Dashed lines indicate possible hydrogen-bond contacts between ring IV, G1517 and U793. (e) In the intermediate RI configuration (green), h24 of the small ribosomal subunit sterically blocks movement of P-site tRNA into the P/E configuration of the fully rotated RF ribosome. Arrows indicate the direction and extent of tRNA and h24 movements during the transition from the intermediate RI configuration to the fully rotated RF configuration.