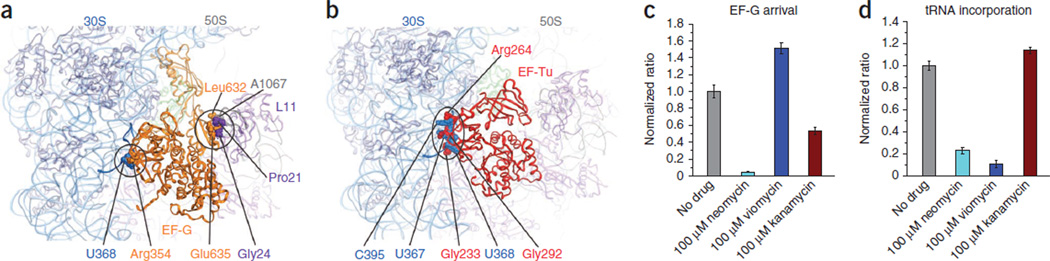
Figure 5.
The intermediate ribosome configuration is incompatible with productive EF-G and EF-Tu binding. (a) The intermediate ribosome structure superimposed on the Thermus thermophilus ribosome structure containing EF-G (gold)48. (b) The intermediate ribosome structure superimposed on the T. thermophilus ribosome structure containing EF-Tu (red)53. In a and b, specific points of steric clash between the elongation factors and the intermediate ribosome configuration are indicated. EF-G domain II clashes with the 30S body near h15 (blue spheres), and EF-G domain V clashes with the L7/L12 stalk protein L11 (purple spheres) and residue A1067 within H43 of 23S rRNA (gray). Domain II of EF-Tu clashes with the 30S body. Superpositions were carried out using the PyMOL ‘pair_fit’ function61. (c) The normalized ratio of individual pre-translocation complexes showing EF-G binding events in the absence of drugs (gray) or in the presence of neomycin (cyan), viomycin (blue) or kanamycin (maroon). All data are shown as the mean ± s.d. from three independent experiments. (d) The normalized ratio of individual empty A-site (after initiation) complexes showing ternary complex binding events in the absence of drugs (gray) or in the presence of neomycin (cyan), viomycin (blue) or kanamycin (maroon). All data are shown as the mean ± s.d. from at least three independent experiments.