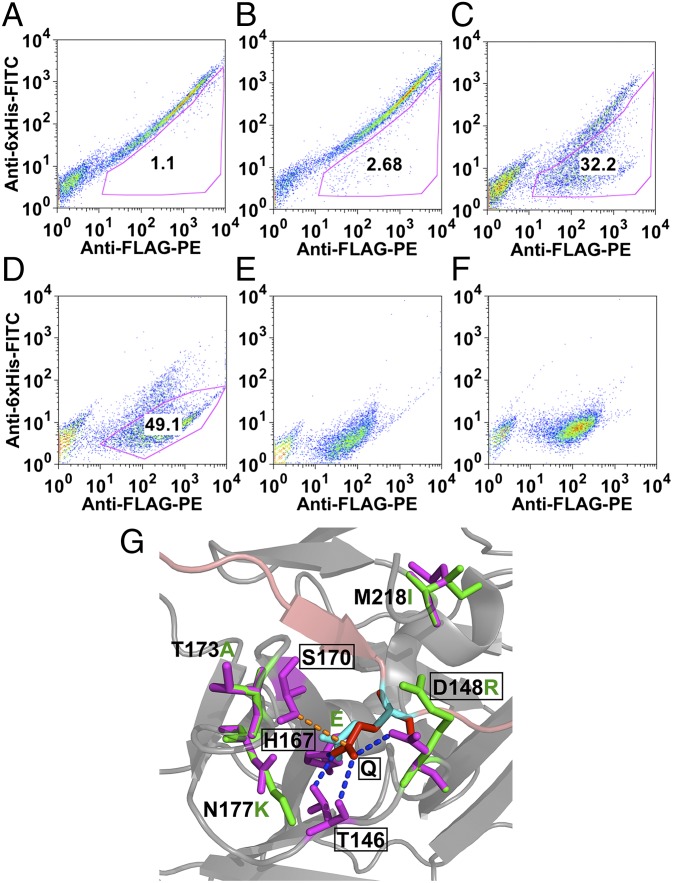
Fig. 3.
FACS analysis of TEV-P S1 subsite library, using the YESS system. (A) Two-color FACS analysis of the library cells. (B–D) Two-color FACS of cells after the first, second, and third round of sorting. (E) The TEV-PE3 variant. (F) The TEV-PH7 variant. (G) Molecular image of the S1 pocket of the wild-type TEV protease and the amino acid substitutions introduced in the TEV-PE10 variant. The P1 residue (Gln, red) of the substrate peptide (pink) interacts with the TEV protease S1 pocket residues (emphasized in rectangular boxes) through the hydrogen bonds (blue dotted line) and the hydrophobic interactions (orange dotted line). In the TEV-PE10, mutations that are close to the new S1 pocket (P1 residue is replaced with Glu, cyan) were annotated (purple in TEV-P and green in TEV-PE10). The image was generated on the basis of the Protein Data Bank file 1LVB, amino acid substitutions were added using PyMol’s mutagenesis function, and no computer simulation was applied.