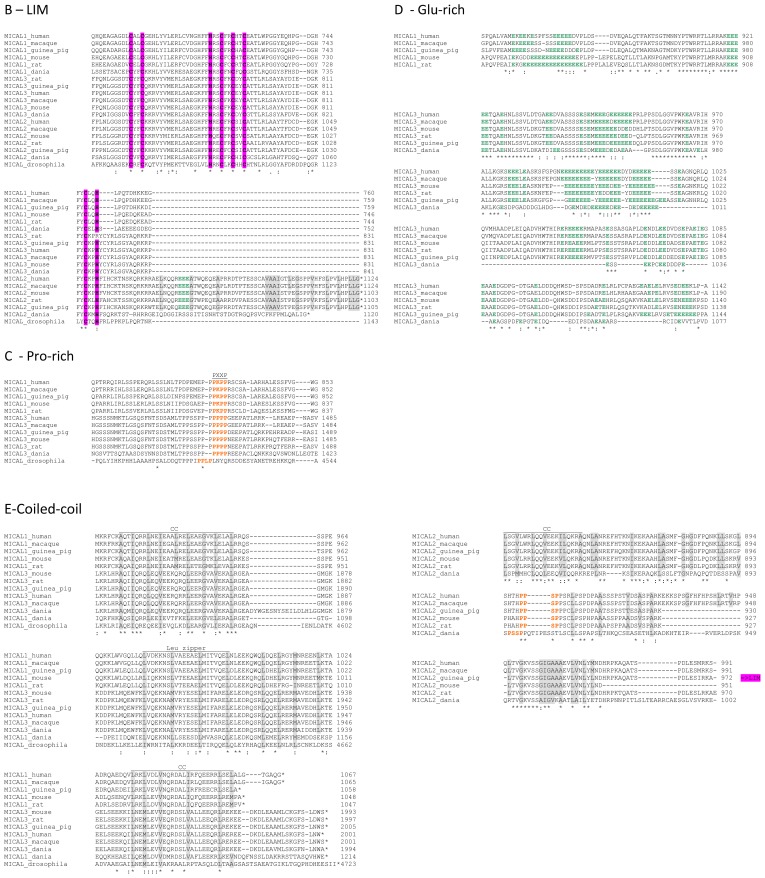
Figure 4.
Sequence comparison of MICAL forms. Regions corresponding to the monooxygenase (MO) and calponin homology (CH) domains (A), LIM domain (B), Pro-rich (C), Glu-rich (D) and C-terminal coiled-coil regions (E) of selected MICAL sequences (Table 1) have been aligned with ClustalW2 [61]. The output reflects the degree of sequence similarity among the proteins. The sequences were annotated as follows, with the numbering of mouse MICAL1. A: cyan, basic residues in the N-terminal proteins regions forming one of the patches of basic potential on MICAL-MO. In bold are conserved residues identified by Nadella et al. [4] and those identified by us from sequence and structural comparisons. Yellow, residues implicated in FAD binding as specified above the sequences. The residues matching the ADP binding region identified as in [62] and those matching the second FAD consensus sequence identified based on [63] are in bold; the additional residues that interact with the isoalloxazine ring (i), the pyrophosphate (p), adenosine (Ado) and AMP regions of FAD, as identified in [3], are also indicated. Red, residues proposed to be involved in the interaction with NADPH as deduced from the comparison with PHBH [64] and structural analysis [3,4]. In bold are residues indicated by [3]. In plain characters are those identified by us as also contributing to the formation of the patch of basic potential for NADPH binding. The conserved regions of MICAL’s MO domain are indicated with the red stars below the aligned sequences as in [3]. In the CH domain, residues forming the hydrophobic core as based on the structure of the CH domain of human MICAL1 (PDB ID 2DK9, [65–67]) are underlined. The regions corresponding to the actin binding site (ABS) of type 2 calponin homology (CH2) domains and to the phosphatidylinositol (4,5)-bisphosphate (PIP2) binding site also found in type 2 CH domains are also indicated. Residues matching the consensus sequence for the formation of the ABS and PIP2 binding sites, as identified in [65,66], are in bold; B: in the LIM domain, the conserved C683, 686, 707, 710, 713, 733 and H704, H736 residues proposed to coordinate the zinc ions in the zinc-finger motifs according to reference [18] are bold and purple. In MICAL2 sequences, the C-terminal protein region next to the LIM domain is included; C: the Pro-rich regions of MICAL1 and 3 matching the PXXP consensus sequence for the formation of a SH3 binding domain initially identified in MICAL by Suzuki et al. [1] are in orange. In MICAL2 a sequence matching the PXXP motif is found before the LIM domain and is shown in orange in panel E; D: the Glu-rich regions found in MICAL1 and 3 are aligned separately with the Glu residues in green. A Glu-rich region is highlighted green in the C-terminal region of MICAL2 in B; E: the regions potentially forming coiled-coils (CC) are aligned for MICAL 1 and 3 and for MICAL2. The hydrophobic residues of the potential heptad repeats are colored grey. The region potentially forming a leucine zipper in MICAL1 and 3, as identified in [1], is indicated. In MICAL2, the LIM domain (B) is after the putative coiled-coil region, and is followed by a region where a coiled-coil structure may also be formed (see B).*, identical residues; :, conserved residues.