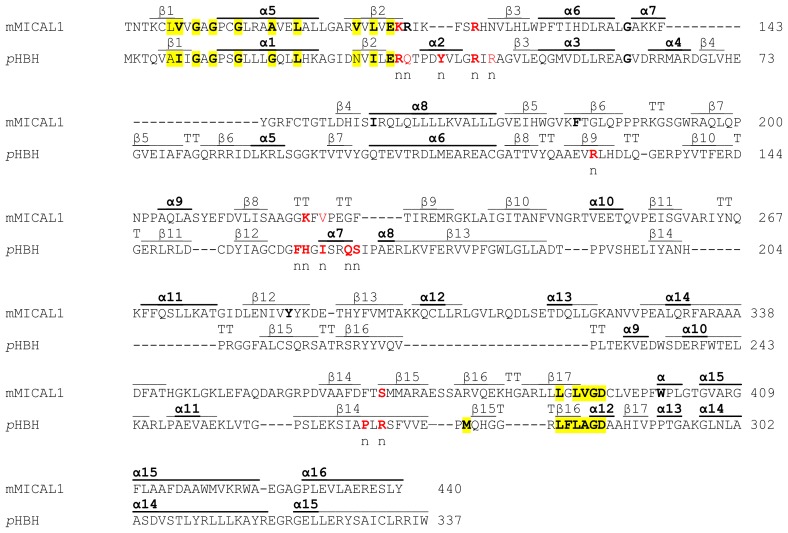
Figure 5.
Comparison between mouse MICAL and p-hydroxybenzoate hydroxylase. The 83–440 region of mouse MICAL1 is compared with full-length Pseudomonas fluorescens p-hydroxybenzoate hydroxylase. The alignment takes into account the known three-dimensional structures of the proteins (2BRA [4], 2BRY [3] for mouse MICAL; 1PBE [68] for PHBH) as in [3,4]. The secondary structure elements (β strands, α helices and β turns (TT) are indicated above each sequence. The conserved residues in the consensus sequence for the formation of the binding site of the adenylate portion of FAD [62] and the second FAD consensus sequence of Eggink et al. [63] are colored yellow. The residues that in PHBH are implicated in NADPH binding [64] are indicated in red and with “n”. If they are conserved in equivalent positions in all MICALs (see Figure 4) they are in red in both sequences. Ser368 (conserved as S or C in all MICAL sequences) proposed to be equivalent to Arg269 of PHBH [3] is also indicated.