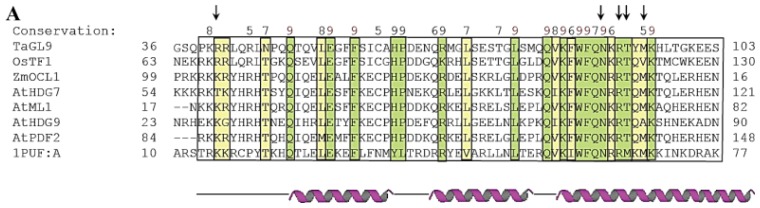
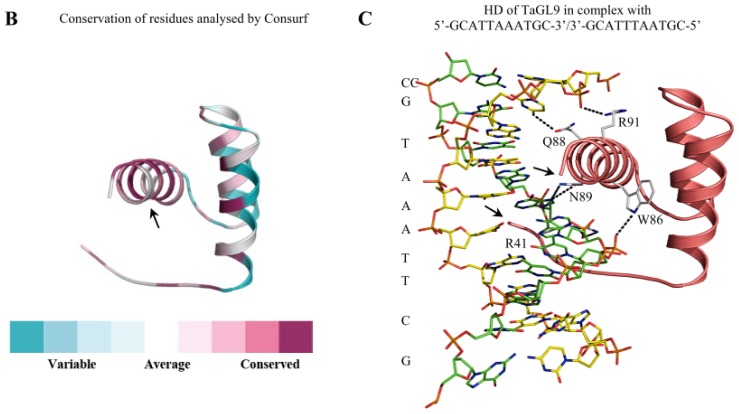
Figure 2.
Molecular modelling of the homeobox domain (HD) of TaGL9 from wheat in complex with an 11-bp long DNA fragment. TaGL9 has at least six domains as predicted by ProDom [56]. (A) A multiple sequence alignment of selected HD sequences using ProMals3D [51]. The predicted secondary structures are shown in magenta (α-helices) and black (loops). Conservation of residues on a scale of 9–5 is shown at the top of the diagram. The absolutely conserved and similar residues are shaded in green and yellow, respectively. The black box indicates the boundaries of HD domains. Vertical arrows above the alignment point to the DNA-interacting residues shown in panel C; (B) HD structure of TaGL9 showing the degree of conservation. The coloured model is based on known HD-Zip IV protein sequences found in the Consurf database [52]. 138 sequences were found by CSI-BLAST [53] but only 66 sequences were unique for the Consurf algorithm to perform calculation. Highly conserved amino acids are coloured in deep magenta, while the least conserved and average ones are coloured in cyan and white, respectively. The black arrow indicates the third helix, for which there is a particularly high level of amino acid conservation; (C) A molecular model of HD of TaGL9 in complex with 5′-GCATTAAATGC-3′/3′-GCATTTAATGC-5′; the model was constructed as described [57]. HD of Hox-A9 from mouse (Protein Data Bank accession 1puf, chain A), in complex with a 20-bp DNA duplex fragment 5′-ACTCTATGATTTACGACGCT-3′ [50] served as a template. Ribbon representation in salmon shows the disposition of secondary structure elements. Here, the α-helix 3 (perpendicular to the viewer’s plane) carries most of the residues that mediate contacts between HD and the DNA fragment. The duplex DNA is shown in cpk-green (coding strand) and cpk-yellow (complementary strand). The nucleotides interacting with HD are represented as cpk sticks. The left and right black arrows point to the NH2- and COOH-termini of HD, respectively. Separations of ≥3.5 Å between the contacting residues (1-letter codes) of HD and a DNA strand are indicated by black dotted lines. The interplay of the interacting residues within HD suggests that structural rigidity and/or flexibility could impact upon selectivity of DNA binding. It is of note that mainly the TAAA and GCAT segments of the coding and complementary strands, respectively, are interacting with the five highly conserved residues of HD. The nucleotide sequence of the DNA coding strand is shown on the left.