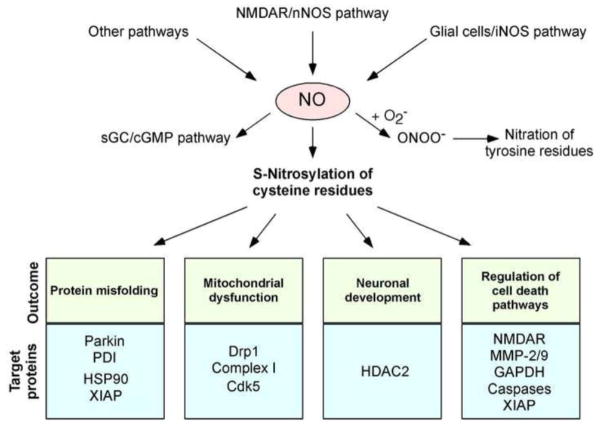
Fig. 1.
Possible mechanisms whereby NO signaling regulates neuronal function. Hyperactivation of neuronal NMDARs can induce activation of neuronal NO synthase (nNOS) and thus production of NO. Glial cells (astrocytes and microglia) can also generate NO via iNOS expression or ROS-dependent inhibition of astrocytic glutamate uptake, which then activates neuronal NMDARs (Akama and Van Eldik, 2000; Lauderback et al., 1999; Li et al., 2009; Weldon et al., 1998). Additional mechanisms, such as mitochondrial dysfunction, may exist to promote NO production in the nervous system. Endothelial cells in the CNS vascular niche may also produce NO. NO thus generated can trigger formation of S-nitrosylated proteins. NO also activates soluble guanylate cyclase (sGC) to produce cGMP, which can activate cGMP-dependent protein kinase. Peroxynitrite (ONOO−), derived from reaction of NO and superoxide anion (O2−·), can nitrate tyrosine residues to form 3-nitrotyrosine. Physiological levels of NO mediate neuroprotective effects, at least in part, by S-nitrosylating the NMDAR and caspases, thus inhibiting their activity. NO can also promote neuronal development via S-nitrosylation of HDAC2. In contrast, we and others have mounted evidence that overproduction of NO can be neurotoxic via S-nitrosylation of parkin, PDI, GAPDH, MMP-2/9, Cdk5, and Drp1. For instance, S-nitrosylated parkin and PDI contribute to neuronal cell injury by triggering accumulation of misfolded proteins, and S-nitrosylation of Drp1 causes excessive mitochondrial fragmentation and thus synaptic damage in neurodegenerative conditions. Additionally, since HSP-90 is a molecular chaperone and XIAP is a ubiquitin E3 ligase, S-nitrosylation of these proteins may contribute to protein misfolding and accumulation in degenerating neurons.