Figure 1.
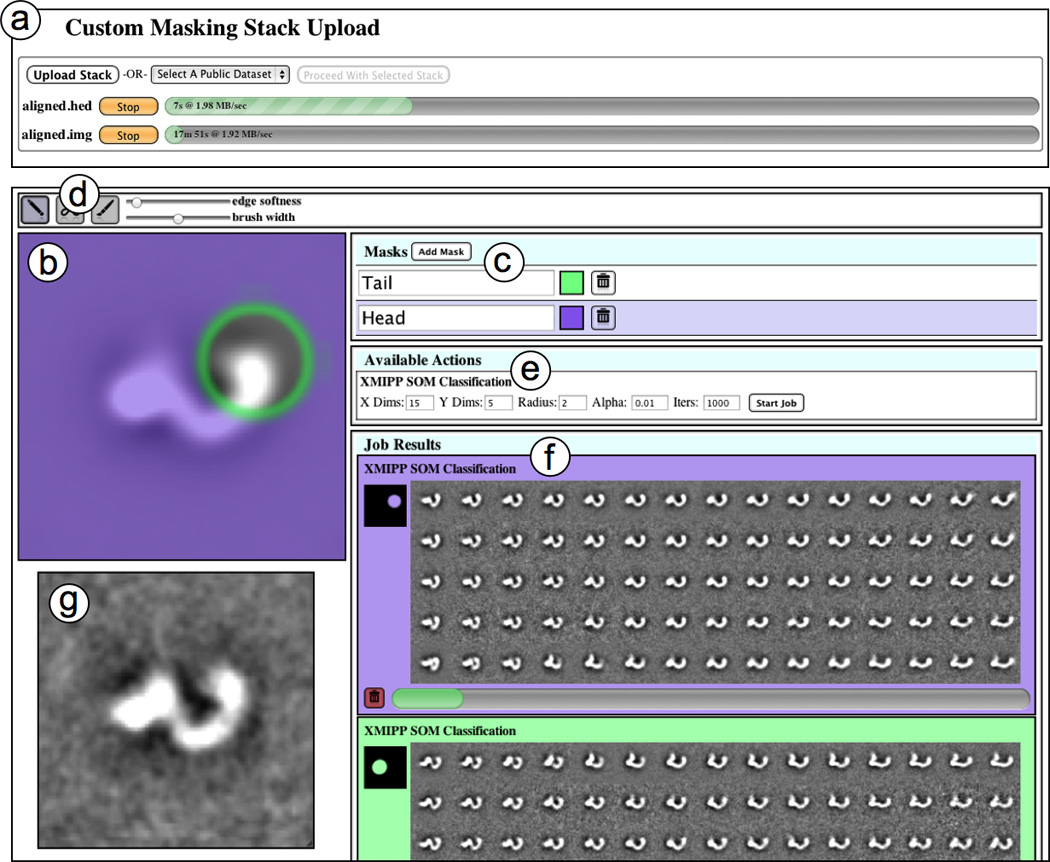
An overview of the Maskiton web interface. This interface allows custom masks to be drawn so that classifications can be focused on specific areas of a dataset. (a) Users can upload new stacks by selecting them on their local machine, or they can select one of the public datasets. Since uploads are resumable, selecting a stack that has already been uploaded is equivalent to selecting the same stack on the server. (b) The stack average is displayed and the mask currently being edited is superimposed on it. The edges of the masked areas are highlighted in green. (c) New masks are created, and can be given labels and colors to keep them organized. The mask superimposed above the stack average is selected by clicking on its entry in the list. (d) Simple drawing tools for editing masks are available. These include a brush, eraser, and sliders for changing width and softness. The softness of mask edges is reflected by the width of the green outline on the mask. (e) The classification parameters are set and new jobs are started on the server by pressing the ‘Start Job’ button. (f) Results for classification jobs started in the current session are displayed, and the results and progress bar are updated as the classification progresses. (g) A zoom-in viewer can be opened by clicking on any of the class images in the results. This viewer displays the class currently beneath the cursor, and allows for movie-like browsing and comparison. This zoom-in view can be moved and resized for convenience.
