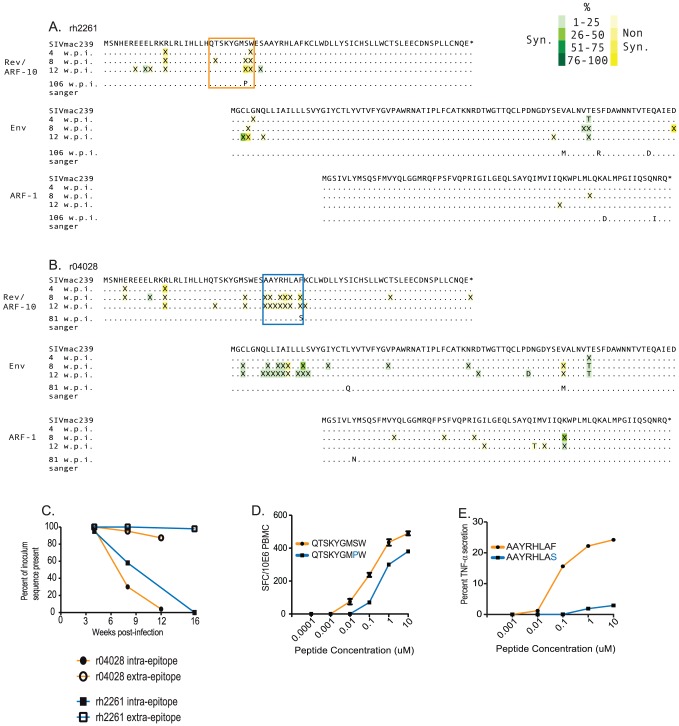
Figure 3. Viral sequence evolution in overlapping reading frames encoding Env and ARFs -1 and -10.
We used next generation pyrosequencing (454 Life Sciences) of the amplicon depicted in figure 1 to sequence the portion of SIV encoding ARF-10, ARF-1 and the first 89 amino acids of Env. A) Kinetics of viral evolution at weeks 4, 8 and 12 from animal rh2261. Mutations synonymous in a given reading frame are boxed in green and non-synonymous changes are boxed in yellow. Matching residues are depicted as “.”. Mutations are depicted as “X” when nucleotide mutations in a given codon could give rise to more than one amino acid. The shade of the box surrounding a mutation represents the frequency of underlying mutations at that codon. The key is in the upper right of the figure. Only mutations present at >1% are shown. The minimal epitope mapped in figure 2 is shown boxed in the same animal-specific color coding as in figure 2; The QW9 epitope for animal rh2261, A, and the AF8 epitope for animal r04028, B. We used simple Sanger sequencing of virus derived from each animal at their individual times of euthanasia to determine the consensus escape patterns in this region after the resolution of the acute phase of infection. C) The frequency of amino acid sequences representing the inoculum within the targeted epitope (intra-epitope, solid shapes) and outside the targeted epitopes (extra-epitope, open shapes) within ARF-10 plotted against weeks post infection. D) IFN-γ ELISPOT assay of recognition of the peptide representing the wild type QW9 sequence (orange) versus the escape variant (orange) by PBMC isolated from animal rh2261 from 4 weeks post infection. E) Intracellular cytokine staining (ICS) assay to measure recognition of a CTL line from r04028 against the wild type AF8 peptide (orange) versus the escape variant (blue). Details of the CTL line are described in the text.