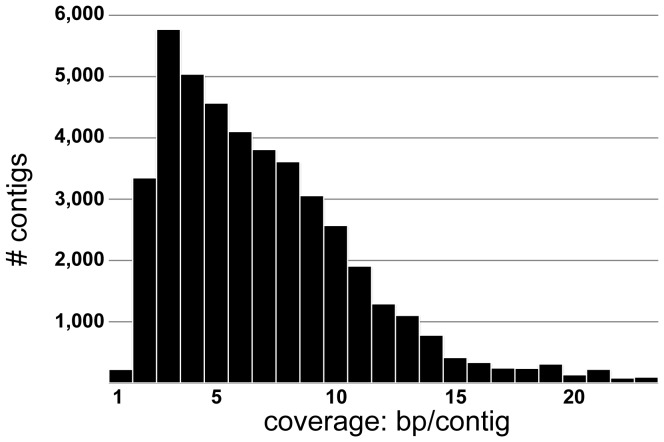
Figure 2. Distribution of average coverage (bp/contig) within contigs produced by de novo assembly of the G. bimaculatus transcriptome.
The coverage within contigs is calculated by dividing the total number of base pairs contained in the reads used to construct a contig by the length of that contig.