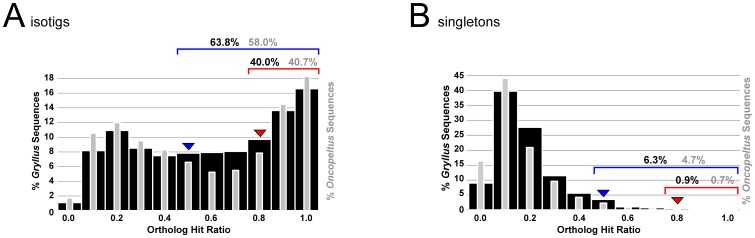
Figure 4. Ortholog hit ratio analysis of the G. bimaculatus de novo assembled transcriptome.
The ortholog hit ratio is a comparison of the length of an assembled sequence to the total length of the full length transcript of its putative ortholog [71]. Values close to one suggest that a transcript predicted by the de novo assembly is close to full length. Ortholog hit ratios for the G. bimaculatus transcriptome sequences are compared to those for the previously reported de novo assembled transcriptome of another insect, the milkweed bug Oncopeltus fasciatus [11]. (A) Ortholog hit ratio analysis of assembled isotigs. A majority (63.8%) of all G. bimaculatus isotigs (black bars) have an ortholog hit ratio of ≥0.5 (blue arrowhead), and 40.0% have an ortholog hit ratio of ≥0.8 (red arrowhead). These values are higher than those obtained for the O. fasciatus de novo assembled transcriptome (grey bars) [11]. (B) Ortholog hit ratio analysis of unassembled singletons. As expected, singletons represent much smaller proportions of putative full-length transcripts. 6.3% of G. bimaculatus singletons (black) have an ortholog hit ratio of ≥0.5 (blue arrowhead), while 0.8% have an ortholog hit ratio of ≥0.8 (red arrowhead). As for the isotig analysis, these values are higher than those obtained for the O. fasciatus de novo assembled transcriptome (grey) [11].