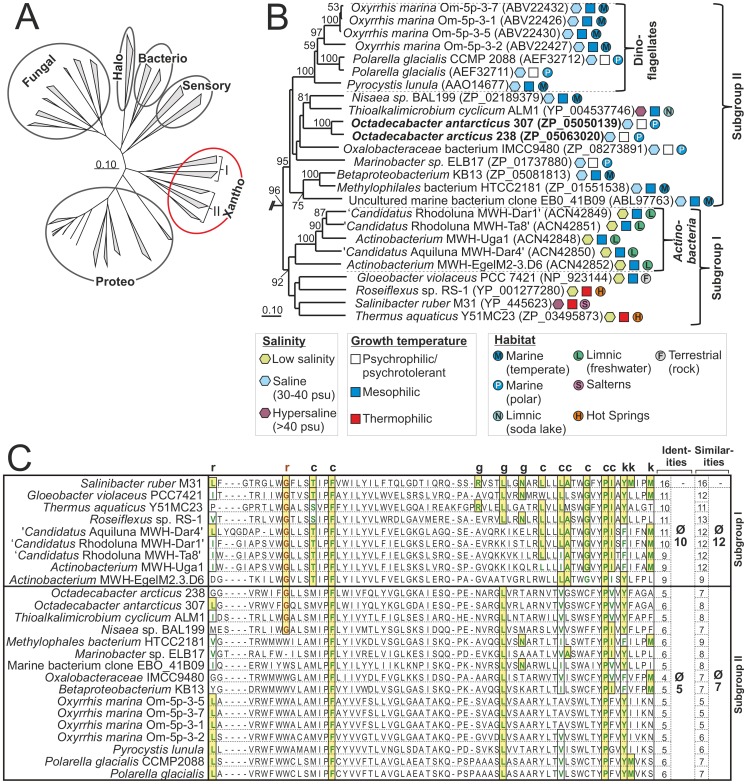
Figure 4. Comparison of the Octadecabacter rhodopsins with other microbial rhodopsins.
(A) Unrooted neighbor-joining tree based on the amino acid sequence of representative members of all known groups of microbial rhodopsins (fungal, sensory, halo-, bacterio-, proteo-, and xanthorhodopsins). The two subgroups of xanthorhodopsins are indicated by the roman letters I and II. (B) Rooted detailed view of the xanthorhodopsin-branch of the same tree. Fungal rhodopsins served as outgroup. Bootstrap-values >50 are given at the respective nodes. The subgroups I and II as well as the Actinobacteria and Dinoflagellate subclusters are indicated by brackets. The lifestyle of the associated organisms is indicated by different symbols. For each rhodopsin the NCBI accession number is given in parentheses. (C) Alignment of the putative keto-carotenoid-binding region of xanthorhodopsins. Residues that interact with the keto-carotenoid in Salinibacter xanthorhodopsin as identified by Imasheva et al. [14] are marked by the letters c, g, k and r, which indicate contact with the chain, glucoside, keto group and ring of the carotenoid, respectively. Similarities are marked by solid boxes and colored letters. Identities are additionally marked by background shading. Amino acids corresponding to Gly156 of Salinibacter xanthorhodopsin are highlighted in red. The number of identities and similarities are given for each xanthorhodopsin and as an average for each subgroup (excluding Salinibacter xanthorhodopsin).