Abstract
The migration of epithelial cells requires coordination of two actin modules at the leading edge: one in the lamellipodium and one in the lamella. How the two modules connect mechanistically to regulate directed edge motion is not understood. Using a combination of live-cell imaging and photoactivation approaches, we demonstrate that the actin network of the lamellipodium evolves spatio-temporally into the lamella. This occurs during the retraction phase of edge motion when myosin II redistributes to the cell edge and condenses the lamellipodial-actin into an arc-like bundle (i.e., actin arc) parallel to the edge. The newly formed actin arc moves rearward and couples to focal adhesions as it enters the lamella. We propose net edge extension occurs by nascent focal adhesions advancing the site at which new actin arcs slow down and form the base of the next protrusion event. The actin arc thus serves as a structural element underlying the temporal and spatial connection between the lamellipodium and lamella to drive directed cell motion.
Introduction
Migrating cells advance by net protrusion at their front and retraction at their rear1. The cell’s leading edge plays a particularly important role in this process through the spatio-temporal control of F-actin, myosin II and focal adhesions, the machinery responsible for cell protrusion2. Two regions define the leading edge: the lamellipodium, a thin sheet of cytoplasm extending ~3-5 μm from the cell edge that consists mostly of dynamic, crisscrossed actin filaments1, 3; and the lamella, the region immediately behind the lamellipodium composed of bundled actin filaments in association with focal adhesions4-6. A major question in the field concerns the interplay between the lamellipodial and lamellar actin modules during cell crawling7-11.
The lamellipodial actin module serves to extend the cell edge. This occurs by insertion of actin monomers into filament ends apposed to the leading membrane and their regulated turnover, whose balance determines the extent of protrusion through actin treadmilling12. The lamellar actin module, on the other hand, assembles a contractile network for traction. This occurs in the lamella through myosin II-based contraction of bundled filaments with arc-like shapes in conjunction with focal adhesions5, 13. Originally, these activities of the lamellipodial and lamellar actin modules were thought to act within one integrated system for driving cell motion, with myosin II working at a distance from the cell edge7. However, in single particle tracking experiments using actin speckling (sptFSM), a small pool of speckles in the lamellipodium was found to have lifetimes and velocities resembling those in the lamella8. These findings gave rise to the view that there was a layer of actin extending through the lamella to the cell edge that controlled forward cell movement14. Known as the “lamella hypothesis,” it envisions that the lamellar actin module plays the primary role in cell crawling, with the lamellipodial actin module subordinate, possibly helping cells to explore their environment in response to extracellular signals15. An elegant version of the lamella hypothesis proposes cell crawling occurs by myosin II contractility in the lamella pulling on the back of the lamellipodium, whose front is tacked down by nascent focal adhesions, resulting in buckling of the lamellipodium and an inchworm-like cell translocation9.
The lamella hypothesis is not without problems, however. Electron microscopy studies show no underlying array of actin that would suggest an extended lamella6, 10. Moreover, long-lived speckles in the lamellipodium that are predicted by the lamella hypothesis have not been detected using alternative speckle tracking tools11.
One obstacle to investigating how the lamellipodium and lamella actin modules connect mechanistically to mediate cell crawling is that the leading edge is both structurally heterogeneous and highly dynamic6, 16. Indeed, there is a shift in the angular distribution of filaments in the lamellipodium during protrusive activity6. This suggests there are dramatic changes in actin organization as the edge undergoes protrusion and retraction on the time scale of minutes. Because maps of sptFSM speckle turnover events typically involve averaging over many protrusion/retraction cycles15 and electron microscopy images of actin distribution provide only a single snapshot of actin organization in time10, how overall actin structure at the leading edge changes to mediate cell movement remains unclear.
Here, we address this question by examining actin turnover with higher temporal and spatial resolution than previously obtained by actin speckle turnover analysis, as well as by examining the overall structural evolution of the actin cytoskeleton over time. We report that the actin network of the lamellipodium evolves into the lamella during the retraction phase of edge motion. This evolution is mediated by myosin II, which redistributes to the cell edge at the beginning of the retraction phase of edge motion, condensing the lamellipodial actin into an actin arc-shaped actin bundle parallel to the edge. We propose the actin arc serves as the structural element underlying the temporal and spatial connection between the lamellipodium and lamella and helps mediate cell crawling through its interactions with focal adhesions.
Results
Actin filament organization at the leading edge examined by EM and FSM
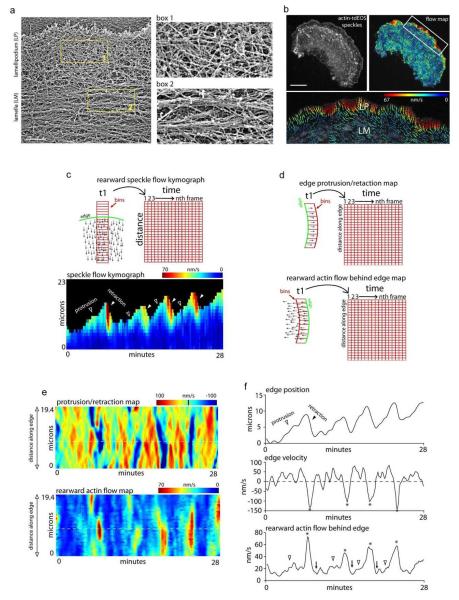
We began by examining the overall structure and dynamics of actin at the leading edge of a migrating epithelial cell using two classic methods, electron microscopy (EM)17 and fluorescence speckle microscopy (FSM)18, relating our findings to prior work5-9, 13, 19, 20. Experiments utilized PtK1 cells because of their large size; their well-spread, flat edges lacking filopodial actin bundles; and their widespread use in previous studies5, 8, 21.
EM of a rotary-shadowed cell showed a criss-crossed actin network in the lamellipodium adjacent to the plasma membrane, and a bundled actin network in the lamella further back (~2-5 μm) (Fig. 1a). There was no evidence of a lamellipodial sheet raised above the lamella as previously reported using this technique in spreading fibroblasts9. Rather, the results were similar to previous ultrastructural findings6, 17, 22, in which crisscrossed actin filaments in the front smoothly transitioned into bundled filaments in the rear with no distinct boundary in between.
Figure 1.
Retrograde actin flow rates change several times over a single edge protrusion/retraction cycle. a) Electron micrograph of a rotary-shadowed cell after live-cell extraction. b) Actin-tdEOS speckle image of an entire PtK1 cell with a corresponding FSM flow map and a higher magnification of the leading edge flow. Vector colors reflect flow speed (color bar), and arrows reflect direction. Scale bars in (a) and (b), 1 μm and 10 μm respectively. c) Edge motion rates relative to retrograde actin flow. Schematic of how the speckle flow data was binned and resulting rearward speckle flow kymograph showing the change in retrograde flow rates during protrusion (open arrows) and retraction (closed arrows) of the leading edge. Each data bin was 5 μm across and 1 μm high. d) Schematic shows how the edge protrusion and retrograde flow data was binned across the leading edge. Bins for edge protrusion were set at 500 nm parallel to the edge. Bins for retrograde flow were set at 1 μm parallel to the edge and 3 μm into the cell. e) Edge protrusion/retraction velocity and rearward actin velocity maps of the same cell used for the kymograph in (c). f) Edge position, edge velocity, and rearward actin flow of the region denoted by the dotted lines in (e) plotted over time. Stars in edge velocity and rearward actin flow graphs denote retractions and arrowheads denote protrusions corresponding to increases in rearward actin flow. Arrowheads denote slowing rearward actin flow immediately after edge retraction.
Rearward actin filament flow at the cell’s leading edge was next monitored by FSM. Actin speckles were created by photoconverting a subpopulation of actin-tdEos molecules from green to red emission in expressing PtK1 cells. Analysis of flow patterns using correlative tracking algorithms23 revealed an overall fast flow zone of actin filaments in the lamellipodium and a slow flow zone in the lamella, as previously reported8, 18(Fig. 1b).
These results demonstrated actin organization in our PtK1 cells resembling that reported in other migrating cells. However, because the EM data represented only a single snapshot of actin structure, while the FSM data represented speckle flow rates at one time point, this data provided limited insight into the complex structural dynamics of actin at the leading edge. This prompted us to employ additional approaches for dissecting the spatially-resolved dynamics of actin at the leading edge.
Actin flow velocity measured during edge protrusion versus retraction
We began by measuring actin flow patterns at high temporal resolution (5 sec intervals) over long periods of time (25-60 min) in order to compare flow rates during protrusion and retraction phases of edge motion. This was accomplished using actin-tdEos to generate actin speckles and a cross-correlative tracking algorithm previously developed to look at actin filament behavior23. Our effective temporal resolution, obtained by averaging three frames acquired at 5-second intervals, was 15 seconds, which provided sufficient resolution to determine whether there were changes in actin flow rates over the cycle of protrusion and retraction underlying edge motion which in on the order of minutes. Previous actin speckling experiments have been limited by photo-bleaching and, thus, many of the published studies using actin speckling focus on edge protrusion. We were able to obtain long time-lapse recordings by replenishing the supply of fluorescently-tagged actin monomers. This was accomplished by converting a small amount of actin-tdEOS to the red channel in the cell body every 40-50 frames. These monomers then slowly incorporated into the actin network at the leading edge.
Using this approach, we generated a kymograph from binned speckle flow vectors showing rearward actin flow velocity during protrusion and retraction of the edge (Fig. 1c diagram). In the cell analyzed in the kymograph of Fig. 1c, the edge undergoes four full protrusion and retraction cycles during the 28 min of imaging. Notably, a large increase in flow velocity always occurs during retraction (see closed arrowheads) (Fig. 1c, kymograph) relative to that observed during protrusion (see open arrowheads).
In a further analysis, we examined the relationship between edge motion and retrograde flow using window sampling of edge protrusion/retraction and rearward actin flow along the entire edge (Fig. 2d diagram-e). This analysis is similar to what has been as previously presented for study of the protrusion phase of edge motion8. Both the protrusion/retraction rate and rearward actin flow patterns were plotted over time in maps colored-coded for speed (Fig. 1e). The retraction phase of edge motion (Fig. 1f, stars in edge motion graph) is always accompanied by a significant increase in the rate of rearward actin filament flow (Fig. 1f, stars), with a smaller increase occurring during edge protrusion (Fig. 1f, open arrowheads).
Figure 2.
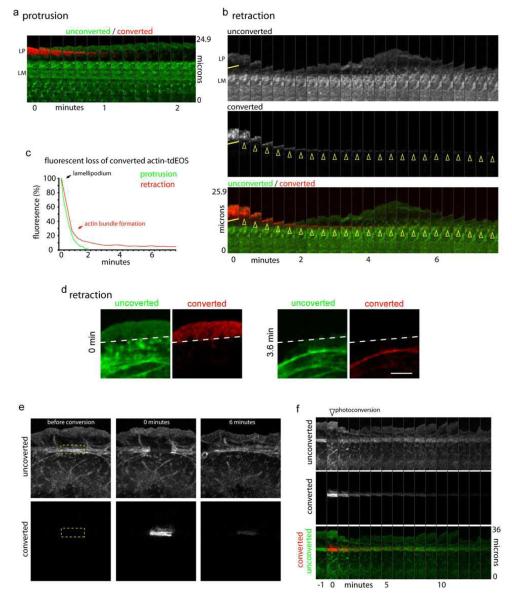
Differential actin filament turnover during protrusion and retraction. a) Montage of unconverted actin-tdEOS (green) at the edge and actin-tdEOS photo-converted in the lamellipodium during edge protrusion. b) Montage of unconverted and converted actin-tdEOS molecules during edge traction. Yellow line denotes region of photo-conversion and arrowheads denote actin bundles formation. c) Quantification of fluorescence loss of converted actin-tdEOS molecules in the lamellipodium during edge protrusion (green line) or retraction (red line). d) Still frames showing the actin bundle formed after retraction is arc shaped. e) Montage showing unconverted and converted channels before, 0 minutes, and 6 minutes after photo-conversion of actin-tdEOS molecules incorporated into actin arcs in the lamella (box). f) Time montage showing the recovery of fluorescence from the unconverted channel and loss of fluorescence from the converted channel of actin-tdEOS in the lamella. Scale bar, 5 μm.
These results were inconsistent with the lamellipodium acting as a simple treadmill machine throughout an edge protrusion/retraction cycle because the flow rate during retraction did not decrease relative to that during protrusion as predicted by treadmilling models8. At the same time, the data did not fit with simple substrate coupling models of edge behavior20, 24, since the flow rate increased during both protrusion and retraction instead of only during retraction. The data instead suggested that as the edge switches between protruding and retracting states, a major change in the lamellipodial actin module occurs, driving it rapidly rearward.
Actin turnover kinetics at the leading edge measured by photoactivation
To gain insight into why actin filaments in the lamellipodium begin to increase in rearward flow rate during the transition from protrusion to retraction, we examined actin filament turnover rates, which are central to maintenance of the lamellipodium actin module11, 15, during edge protrusion and retraction. The necessary temporal resolution was achieved by photoconverting a subpopulation of actin-tdEOS in the lamellipodium of a cell as it underwent either protrusion or retraction (Fig. 2a-d). Because this subpopulation was now spatially highlighted, we could specifically track the fate and lifetime of these actin filaments.
During edge protrusion, photoconverted actin-tdEos molecules in the lamellipodium completely turned over within 2 min, with few, if any, actin monomers incorporated into filaments transferred into the lamella (Fig. 2 a,c). This turnover occurred during rearward flow of the actin network, presumably through filament depolymerization. There was no evidence of a lamellar actin pool with a longer half-life underlying the lamellipodial actin, as envisioned in the lamella hypothesis. Instead, the data indicated that during protrusion the lamellipodial and lamellar actin modules are distinct.
During edge retraction, a subset of photoconverted actin-tdEos molecules persisted within actin filaments (see yellow arrowheads in Fig. 2b, graph in Fig. 2c, and Fig. 2d). These compacted into an arc-shaped bundle that moved rearward with retrograde flow into the lamellar region. The actin arc then joined the transverse actin filament bundles below it that were not photoconverted (Fig. 2d). These data indicated that as the edge switches from protruding to retracting, a major structural change in the lamellipodium actin occurs, in which it converts into an actin arc. Furthermore, the fast rearward movement of the arc is consistent with the speckle data obtained above showing increased actin flow during edge retraction.
When we photoconverted actin-tdEOS molecules in the lamella, composed predominantly of stacked arcs, the signal remains for only 6-8 min, with most of it lost within the first 2 min (Fig. 2e, f). The kinetics of signal loss resembled that seen for photoconverted molecules in the retracting lamellipodium (see Fig. 2b). Signal loss did not appear to be due to arc disassembly because unconverted signal fills in the area of converted signal quickly with no overall change in the arc stack architecture (Fig. 2e, f). This suggested that even though the arc stack of the lamella has a relatively stable appearance, it is highly dynamic, with actin molecules continually dissociating from and re-associating with it.
Characterization of actin arc formation and behavior
To better understand actin arc formation and dynamics, we performed time-lapse imaging of actin-mRFP in live cells without speckling or photoactivation (Fig. 3). This allowed us to monitor the behavior of actin arcs in relation to other actin structures at the leading edge. These included the actin filament networks in the lamellipodium (Fig. 3a, LP) and the perpendicular actin filaments associated with focal adhesions13 (Fig. 3a, yellow arrowheads). We found that a new actin arc forms in the lamellipodium during every edge retraction (Fig. 3a-c, red arrowheads, Movie S2). The new actin arc moves in a rearward fashion and becomes part of the existing actin arc population in the lamella during the next protrusion phase (Fig. 3a-b, red arrowheads). Time-lapse imaging of cells co-expressing the focal adhesion protein, zyxin, and actin revealed that actin arc bundles can form prior co-localization with focal adhesion sites (Fig. 3d).
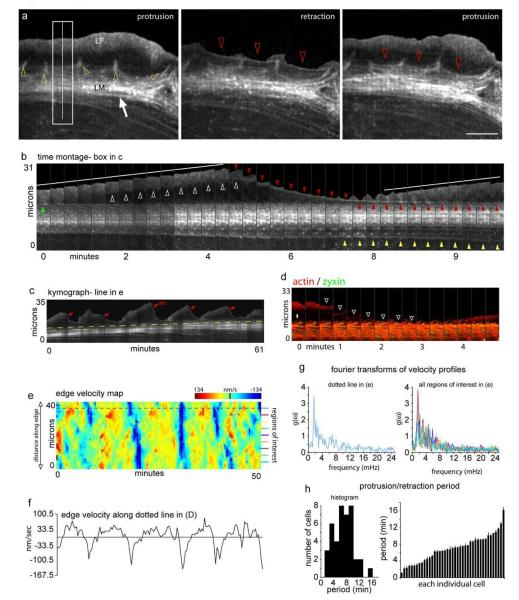
Figure 3.
Actin arc dynamics at the leading edge. a) Three frames of a time-lapse recording of actin-mRFP showing the cytoskeletal organization at the leading edge during the transition from protrusion to retraction and back to protrusion. Star denotes actin network in the lamellipodium and arrow denotes actin arcs in the lamella. Yellow arrowheads show actin filaments associated with focal adhesions. Red arrowheads show a newly forming actin arc. b) Time-lapse montage of the box in (a) showing the formation of an actin arc (red arrowheads) between protrusion events (white lines). Green arrowhead shows the actin arc formed during previous retraction event. Yellow arrowheads show the removal of actin arcs. c) Kymograph of the line in (a) showing multiple protrusion and retraction events over 1 hr. Red arrows denote the first frame an actin arc was observable and yellow dotted line shows lamellar advance. d) Actin-mGFP and zyxin-mCherry montage showing an actin arc can form (white arrowheads) before coming in contact with focal adhesions (yellow arrow). e) Protrusion/retraction map showing edge velocity (shown by color bar) changes over time across the edge of the cell in (a). F) Plot of the edge velocity from the dotted line in (e). g) Fourier transforms of the velocity profiles represented by the colored lines in (e). h) Distribution of the period of the protrusion/retraction cycle among 41 cells. Scale bar, 10 μm. The change in intensity in (b) from frame 10 to 11 is due to focusing.
The velocity of an actin arc decreases when it comes in contact with focal adhesions (Fig. 3d, arrowheads), possibly due to known coupling between actin filaments and focal adhesion proteins25, 26. Consistent with this, we found the rearward motion of regions of an actin arc nearest to focal adhesions to be slower than that of regions furthest from focal adhesions. This creates a bowed appearance in the arc as it enters the lamella (Fig. 3a, red arrowheads). Rarely, a small actin bundle is left behind by the protruding lamellipodium (Fig. S1). This bundle stays relatively stationary until it merges with the retracting primary actin arc (Fig. S1). In the lamella, new actin arc arrival at the front of the arc stack is balanced by actin arc loss and disassembly at the rear (Fig. 3b, c, see red and yellow arrowheads). This results in a treadmilling motion of arcs through the stack, with arcs periodically dissociating from the back of the lamella and disappearing (Fig. 3b, yellow arrowheads, Movie S2).
Velocity profiles of the edge over time were analyzed in protrusion/retraction maps (color-coded for velocity), as well as in graphs that displayed single points along the edge (Fig. 3 e, f). This revealed that the frequency of protrusion and retraction of the cell edge (and hence frequency of arc formation) remains constant over time. Fourier analysis to determine the variation in the velocity with time at each point along the edge of an individual cell revealed a primary frequency of fluctuation of edge velocity (Fig. 3g), indicating the entire edge of a cell exhibits the same protrusion/retraction frequency. Protrusion/retraction periods analyzed in this manner for many individual cells (n=41 cells) revealed each cell has a characteristic protrusion/retraction period for edge motion (and hence arc formation rate), with the value varying considerably from cell to cell (Fig. 3h).
Role of Myosin II in actin arc formation
Given the above characteristics of actin arcs, we next investigated what mediates their formation. Mature actin arcs in the lamella are myosin II-contractile bundles and inhibition of myosin II activity decreases the number of arcs in both the central domain of neuronal growth cones and the lamella of epithelial cells13, 27. These observations led us to investigate a potential role of myosin II in arc formation. Cells co-expressing myosin IIA-GFP and actin-mRFP were monitored by time-lapse imaging. The changes in distributions of these molecules were specifically followed during protrusion and retraction phases of edge motion.
During edge protrusion, most of myosin II is associated with actin arcs of the lamella, with virtually none found in the lamellipodium (Fig. 4a). This finding supports previous work suggesting that myosin II is restricted to the lamella and is absent from the lamellipodium5, 7, 28. We next monitored myosin II over a protrusion/retraction cycle. This is illustrated in Fig. 4b, which shows a time-montage of the rectangular area of the cell in Fig. 4a. Notably, myosin IIA appears in the lamellipodium at the peak of protrusion (Fig. 4c, white arrowheads). It then translocates backward, mirroring the position and movement of the retracting actin arc (Fig 4c, white arrows). Myosin II also appears on the smaller actin arcs that are occasionally left behind by the protruding lamellipodium (Fig. 4c, yellow arrowhead). This myosin II population remains stationary and then moves rearward with the primary actin arc during edge retraction (Fig 4c, yellow arrowheads). Myosin II thus localizes to actin arcs formed in the lamellipodium during edge retraction.
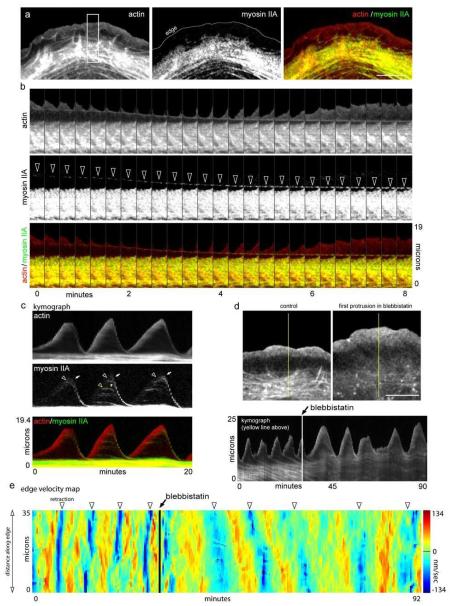
Figure 4.
Myosin II activity condenses the lamellipodium into an actin arc. a) Organization of actin-mRFP, Myosin IIA-GFP, and overlay during edge protrusion. Notice myosin II localizes with older actin arcs in the lamella. b) Time-lapse montages of actin-mRFP, myosin IIA-GFP and an overlay showing the co-localization of myosin II with newly forming actin arcs. Arrowheads show co-translocation of myosin IIA and the newly formed actin arc. c) Kymograph showing myosin IIA dynamics over three protrusion/retraction cycles. Arrowheads denote the appearance and arrows denote the movement of myosin IIA. d) Actin-mRFP before and after 25 μm blebbistatin. Kymograph shows the protrusion retraction cycle of the edge before and after blebbistatin. Note that the structure and movement of the actin arcs is diminished in the presence of blebbistatin. e) Protrusion/retraction map showing edge motion before and after blebbistatin addition (arrow). Edge retractions denoted by arrowheads. Scale bars, 10 μm.
To test whether myosin II is involved in arc formation, we imaged cells treated with blebbistatin, a specific inhibitor of myosin II ATPase activity (Fig. 4d). After treatment, no actin arcs formed and stacked arcs in the lamella disappeared. Indeed, the boundary between lamellipodial and lamellar actin modules was lost in treated cells. This suggested that myosin II activity is required for arcs to form and be maintained, both in the lamellipodium and lamella. Interestingly, the edge protrudes out further in blebbistatin compared to control before retracting. This suggests that myosin II-based actin arc formation regulates but is not the sole controlling element in the switch between edge protrusion and retraction.
Relating edge protrusion/retraction to cell crawling
Previous studies have speculated that the frequency of the edge’s protrusion/retraction cycle at the leading edge is related to how fast the edge exhibits net extension7. This predicts that the diversity in protrusion/retraction periods observed in a cell population (see Fig. 3H) results from differences in crawling rates. We tested this prediction.
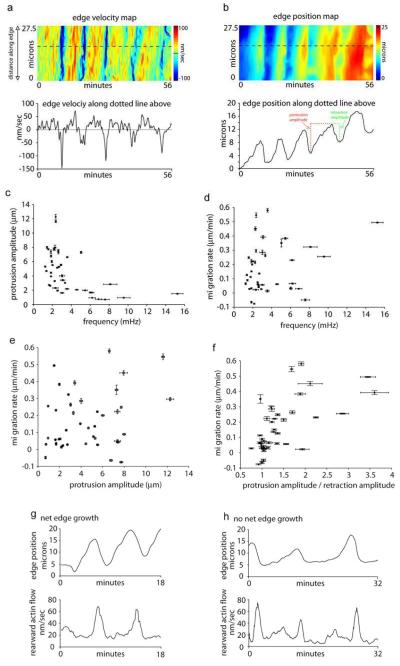
Edge velocity and position data (Fig. 5a-b) in crawling cells allowed us to compare the frequency of edge protrusion/retraction and cell crawling rate (Fig. 5c-f). As expected, an inverse relationship exists between the amplitude and frequency of edge protrusions since it takes a longer time to move the cell forward in cells with larger protrusions vs. smaller protrusions (Fig. 5c). However, we found no relationship existed between the migration rate of a cell and either the amplitude, or frequency, of its protrusions (Fig. 5d-e). Indeed, both the frequency and amplitude of protrusion in an individual cell remains constant, whether or not the cell is crawling. Instead, the major, and obvious, difference in edge motion between fast and slow moving cells is the ratio between the amplitude of protrusion and amplitude of retraction (Fig. 5f). Cells displaying net movement have retraction amplitudes that are smaller than protrusion amplitudes, whereas those that do not move forward efficiently have amplitudes of protrusion and retraction that are roughly the same. This is shown graphically in the colored amplitude measurements of Fig. 5b (see red and green dotted lines). Note that for this particular cell the amplitude and frequency throughout several protrusion/retraction cycles remains constant. Despite this, during the first 20 min the cell underwent little net edge movement, whereas in the last 20 min there is significant movement. This suggests that differences in actin dynamics per se do not explain differences in migration rates. Consistent with this, crawling and non-crawling cells displayed similar patterns of rearward actin flow (Fig. 5g-h). This led us to investigate what features of leading edge dynamics are relevant for whether the edge migrates forward or remains stationary.
Figure 5.
Oscillatory edge motion and net edge extension. a) Edge velocity map along the edge and a single region (bottom graph) along the edge from a crawling cell. b) Edge position map of the same cell as in (a). Edge position map was created by color-coded the lowest edge position as blue and the highest as red as in the color bar. This allows for relative edge position along the same regions as in the velocity map in (a) to be graphically displayed over time. Bottom graph in (b) shows the relative edge position at one point along the edge. Red dotted line and green dotted line graphically show the protrusion amplitude and retraction amplitude of one protrusion respectively. c) Protrusion amplitudes plotted against edge oscillation frequencies for individual cells. Correlation coefficient: −0.5129 (confidence interval: −0.7087, −0.2437). d) Migration rate plotted edge oscillation frequencies for individual cells. Correlation coefficient: 0.1966 (confidence interval: −0.1182, 0.4755). e) Migration rate plotted against protrusion amplitudes for individual cells. Correlation coefficient: 0.2650. (confidence interval: −0.0464, 0.5295). f) Migration rate plotted against the ratio of protrusion and retraction amplitudes in individual cells. Correlation coefficient: 0.4254 (confidence interval: 0.1355, 0.6482). g-h) Edge position and rearward flow velocity plotted as in Fig. 1(f) for a cell that demonstrates net edge growth (g) and a cell that does not (h). Note the pattern of rearward actin flow is similar in both cells. Pearson’s correlation coefficient was used to quantify the correlation and the 95% confidence interval for each pair was computed by using the Fisher transformation.
Defining the protrusion base in crawling cells
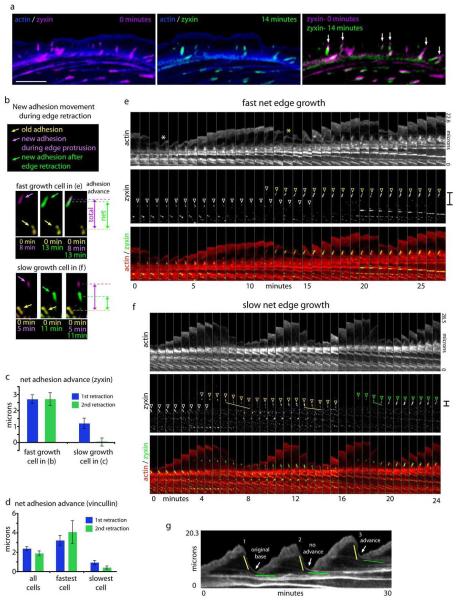
New focal adhesion placement in front of old adhesions is a hallmark of crawling cells and appears to advance the boundary between the lamellipodium and lamella29. Recent work has also indicated that whole adhesions can move away from the leading edge30. To investigate whether this is important for whether the edge migrates forward, we examined the dynamics of focal adhesions in relation to net edge motion. We started by confirming that focal adhesion plaques can move away from the edge. Figure 6a shows focal adhesions labeled with zyxin-mCherry before (purple) and after 14 minutes (green). Note that several of the adhesions moved away from the edge over this time period (Fig. 6a, arrows). We tracked adhesions labeled with either zyxin or vinculin over time and found the rearward movement of focal adhesions occurs coincident with edge retraction (Fig. S2).
Figure 6.
Differential slippage of focal adhesions in crawling vs. non-crawling cells correlates with new actin arc movement. a) Overlay of actin-mGFP (blue) and zyxin-mCherry either at 0 minutes (magenta) or after 14 minutes (green). Arrows in zyxin-mCherry overlay show focal adhesions that move during this time period. b) Focal adhesion movement compared to preexisting adhesions. New focal adhesions during protrusion are labeled purple and the same adhesions after the next edge retraction are labeled green in two cells with different migrating rates (fast cell- 0.25 μm/min; slow cell- 0.04 μm/min). The preexisting adhesion in each cell is diplayed in yellow. Total distance of the new adhesion from the previous adhesion during protrusion is shown by magenta double-headed arrows and the net distance after new adhesion slippage during edge retraction is shown by the green double-headed arrows. c) Quantification net distance between new adhesions and preexisting adhesions for the fast cell in (b) (n= 38 adhesions) and a slow cell in (b) (n= 29 adhesions). This distance was calculated for each new adhesion after the first and second edge retraction event for which they are associated. d) Quanitification of net adhesion advance from focal adhesion labeled with vincullin. Average distance of a population of 8 cells (n= 132 adhesions), and for the fastest (0.31 μm/min; n= 17 adhesions) and slowest (0.01 μm/min; n= 39 adhesions) cell in the population are displayed. e) Time-lapse montage of actin-mGFP and Zyxin-mCherry in a crawling cell. White arrow denotes the base of the first retraction to protrusion transition point and the yellow arrow denotes the second. White arrowhead shows the pre-existing focal adhesion. Yellow arrowhead shows a nascent adhesion appearing and maturing (growing larger). f) Similar time-lapse montage as in (e) in a cell that did not exhibit net forward movement of its edge. White arrowhead denotes a preexisting adhesion. Yellow and green arrowheads denote nascent focal adhesion formation and maturation. Note that adhesion move rearward (lines) during edge retraction. Red and green brackets show the net movement of the base of the protrusion retraction cycle and focal adhesion advance. g) Kymograph from a cell that increases its rate of migration. There is no advance of the base protrusion after two protrusion/retraction cycles (arrows) but there is advance after the third cycle (arrow). Yellow and green lines denote rapid and slow actin arc translocation respectively. Error bars represent standard error of the mean. Scale bar, 10 μm.
We next wanted to define the difference between focal adhesion dynamics in cells that were crawling fast versus slow. Figure 6b shows focal adhesion positions color-coded for time from a cell that is crawling fast compared to one that is crawling relatively slowly. Note that in both cells new adhesions (purple) appear during edge protrusion and are placed at a similar distance from the preexisting adhesions (yellow). During edge retraction, both of the new adhesions (green) move away from the leading edge toward the preexisting adhesion. In the slow crawling cell the new focal adhesion moves a greater distance towards the preexisting adhesion. We measured the net advance of all of the new adhesions in each cell and found that the net advance in the fast migrating cell to be significantly greater than the slow cell (Fig. 6c). The same relationship is observed when focal adhesions are labeled with vincullin-mCherry (Fig. 6d). This focal adhesion slippage can also be observed in focal adhesions labeled with paxillin (Fig. S3).
Since new actin arcs slow down at focal adhesions (Fig. 3d), we next investigated whether these actin arcs interact with newly formed, moving, focal adhesions. The white asterisk in the actin montage of Fig. 6b shows the base of the first retraction/protrusion cycle in the fast crawling cell. This position correlates with where a newly formed actin arc slows down at a pre-existing adhesion (white arrowheads and white asterisk) and a subsequent new protrusion event starts. During this protrusion event, a new focal adhesion is formed (yellow arrowheads). The newly formed actin arc slows at the new adhesion and this becomes the base of the next edge protrusion phase (Fig. 6e, yellow asterisk). This base is positioned distally compared to the first protrusion (compare white and yellow asterisks). These results suggest that new focal adhesion formation helps to advance the cell by slowing down newly formed actin arcs distally to where older actin arcs were slowed during previous edge retractions.
However, in cells that had little net edge extension, new actin arcs move with new adhesions rearward before slowing down together (6f, left yellow and right green lines). Interestingly, the same adhesion moves back further after association with the actin arc from the next retraction even (Fig. 6f, middle yellow line). The net result is that, even over three full protrusion/retraction events, the cell in Fig. 6f advances the base of its protrusion retraction cycle little compared to the cell in Fig. 6e (brackets). We further quantified the additional net adhesion movement after a second retraction event and found that slow crawling cells exhibited a further decrease when the fast cells did not (Fig. 6c-d).
Taken together, our data would suggest that it is the transition between initial rapid actin arc translocation and the slow translocation after coupling to the substrate through focal adhesions that defines where the next protrusion will begin. Interestingly, analysis of actin dynamics from a cell which spontaneously started crawling revealed that the rate of advance of the transition between fast and slow actin arc translocation can be changed within a single cell (Fig. 6g, arrows).
Thus, the fundamental difference between cells crawling rapidly or slowly is not where or when actin arcs form, but the strength of coupling between actin arcs, focal adhesions and the substrate. Since the actin arcs and focal adhesion stay associated with each other in both crawling and non-crawling cells through the subsequent protrusion phase, we propose that the slippage occurs between the focal adhesion and the growth substrate.
The advance of the lamella results from an actin arc treadmill
In order for the protrusion base of the leading edge to continually move forward in the manner described above, coordinated movement of the lamella itself must occur, otherwise the cell would develop too many adhesions to the substrate and stop moving. To clarify how this occurs, we analyzed the spatio-dynamics of actin arcs, focal adhesions and the structural features of the leading edge in a crawling cell.
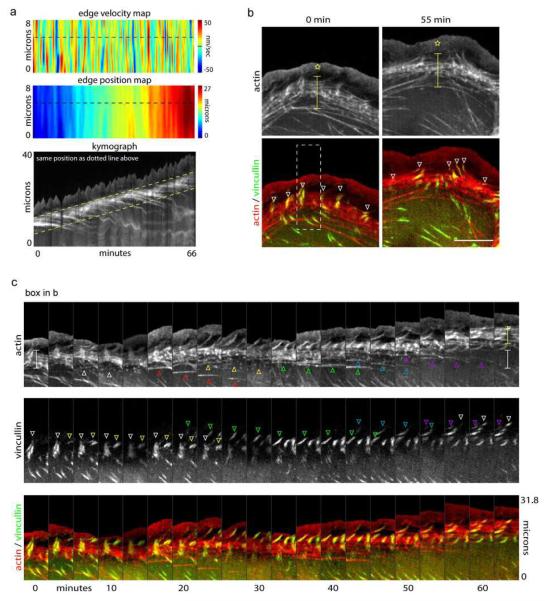
Fig. 7a shows the edge dynamics and actin organization in a crawling cell imaged for 66 min. The edge velocity map (top panel) shows pulsating forward and reverse edge movements. These correspond to the oscillatory protrusion and retraction cycles of the leading edge, as shown in the kymograph in the bottom panel. Two features of the kymograph are notable besides the saw-tooth leading edge pattern arising from the protrusion/retraction cycles. First, the boundary between the lamella and lamellipodium (see upper dashed line in kymograph) moves forward at the same rate as the overall edge, consistent with prior work9, 29. Second, the lamella (representing the zone between the yellow dashed lines in kymograph) maintains a constant width throughout movement.
Figure 7.
The lamella moves forward through actin arc treadmilling. a) Edge velocity map, edge position map and kymograph showing dynamics and advance of the leading edge. Dotted yellow lines denote advance of the lamella. b-c) The lamella advances by treadmilling of actin arcs: b) Actin-mGFP and vincullin-mCherry in a cell before and after net edge extension. Stars denote the lamellipodium and brackets denote the lamella. White arrowheads show focal adhesions at the boundary between the lamellipodium and lamella. c) Actin montage of box in (b) shows actin arcs are removed from the back of the lamella (arrowheads in actin montage) while new focal adhesion assembly (arrowheads in vincullin montage) leads to edge advance. Scale bar, 10 μm. The change in intensity in 5C from frame 5 to 6 is due to focusing.
The first feature of the kymograph, coordinated movement of lamellipodial/lamellar boundary and edge, can be explained by new focal adhesion formation slowing down newly formed actin arcs distally to where older actin arcs are slowed. Since this process determines the base of the next protrusion, the lamellipodial/lamellar boundary and edge undergo coordinated movement. Consistent with this view, focal adhesions were enriched at the boundary between the lamellipodium and lamella. Images of crawling cells co-expressing actin and the focal adhesion protein, vincullin, (Fig. 7b), revealed focal adhesions mainly localize to the boundary between the lamellipodium and lamella at both the early and later time points. Note that in these cells both the lamellipodium (star) and the actin arc stack in the lamella (bracket) maintain their overall width despite considerable edge advancement during the period of imaging.
To explain the second feature of the kymograph in Fig. 7a, maintenance of the constant width of the lamella, we analyzed time montages of actin and vinculin channels obtained from time-lapse images over an extended imaging period (i.e., 66 min). New focal adhesions are placed distally to old ones as the lamellipodial/lamellar interface advances (Fig. 7c, see yellow and green arrowheads), consistent with our findings in Fig. 6 and with previous studies29. Interestingly, once a focal adhesion is placed, both the focal adhesion (see downward pointing arrowheads in vincullin channel of Fig. 7c) and its associated arcs (see upward pointing arrowheads in actin channel of Fig. 7c) disassemble with time. This leads to a treadmill cycle in which new arcs and focal adhesions are added at the front end of the arc stack and disassembled at the rear. Because the rate of arc/focal adhesion entry at the front occurs at the same rate as disassembly at the rear, the lamella maintains a constant width over time. This may explain how cells avoid developing too many adhesive interactions with the substrate, impeding movement, during cell crawling.
Discussion
In this paper we used advanced imaging techniques to clarify the relationship between the lamellipodial and lamellar actin filament modules at the cell’s leading edge. The resulting data supports a new model for leading edge movement and directed cell crawling involving the actin arc.
We began our study by selectively photoconverting tdEOS-labeled actin filaments in the lamellipodium and following their fate over time. The results were difficult to explain using prior models of lamellipodial and lamellar organization, which assumed the two actin modules either are distinct but overlapping networks8, 9, 15, or are one continuous network6, 11. Depending on whether the edge was protruding or retracting, the behavior of the lamellipodial actin filaments differed (Fig 2). During protrusion, labeled actin filaments quickly turned over with none reaching the lamella. During retraction, by contrast, converted filaments transferred to the lamella.
Live cell imaging without photoconversion, using actin-mRFP, helped explain the photoconversion data (Fig. 3). It revealed that during retraction, the criss-crossed actin networks of the lamellipodium undergo rapid conversion into rearward-moving actin bundles or arcs. The arc forms at the cell edge soon after retraction is initiated. It then moves rearward, parallel to the edge, collecting with arcs in the lamella that are compiled as a stack. The presence of arcs as long-lived filaments in the lamellipodium provides an explanation for the differences in lifetimes of photoconverted tdEOS-labeled actin filaments in the lamellipodium, and also explains previous sptFSM data showing long-lived speckles in the lamellipodium8. But the data also raised questions, particularly because previous descriptions of arcs suggest they formed away from the edge in the lamella16, 19, and in response to the presence of focal adhesions31. Our data, however, showed that the stack of arc bundles in the lamella derive from individual arcs formed in the lamellipodium (Fig. 2). Indeed, we observed that some individual arcs form in the lamellipodium prior to co-localization with focal adhesions (Fig. 3). This does not preclude the possibility, however, that forces exerted on the actin filament cytoskeleton by focal adhesions play a role in arc formation31.
These results led us to question what drives the conversion of lamellipodial actin filaments into a rearward-moving actin arc that gives rise to the lamella. An obvious candidate was myosin II16, 27. However, previous reports of myosin II distribution at the leading edge found myosin II restricted to the lamella7, 8, leading to models in which myosin II only modulates actin dynamics in the lamellipodium from a distance9. Since these studies did not take into account potential differences in myosin II distribution when the leading edge was retracting or protruding, we wondered whether the prior studies might have missed a lamellipodial distribution of myosin II. When we performed high resolution imaging of myosin II-GFP dynamics throughout the edge protrusion/retraction cycle, we found that myosin II filaments do, in fact, form in the lamellipodium (Fig. 4). This occurs at the peak of the protrusion phase. The myosin filaments then move rearward with the newly forming actin arc during edge retraction.
The presence of myosin II in the lamellipodium raised the possibility that a local network contraction, similar to that proposed to drive the cell body in migrating fish keratocytes17, transforms the criss-crossed actin filaments in the lamellipodium into the bundled filaments that constitute the actin arc. Linked to the plasma membrane, this conversion could be the driving force of edge retraction seen in migrating cells. Supporting this idea, no actin arcs form and the edge protrudes further relative to that in control cells when myosin II activity is suppressed (Fig. 4).
Given the finding that lamellipodial actin converts into an arc parallel to the edge, and subsequently moves rearward into the lamella, we asked how this relates mechanistically to net edge extension and cell crawling. A clue came from our finding that the amplitude of the protrusion phase of the cell’s oscillatory edge cycle remains constant and does not change even if the cell migrates forward. This means that net forward edge extension requires a process that shifts the base where the protrusion phase initiates. We found that the base where the protrusion phase initiates is the zone where the arc slows down upon moving into the lamella. This zone contains focal adhesions, which act as a brake on the rearward moving arcs, slowing them down as they enter the lamella. This results in a single actin arc spanning many focal adhesions across the edge. Stress fibers have recently been shown to mechanically link multiple large focal adhesions across the cell body in non-crawling cells32. Therefore, a linked actin arc/focal adhesion network at the leading edge could act during edge protrusion as a coherent stiff substrate for actin filaments in the lamellipodium to push back against to extend the plasma membrane.
Based on these results, we propose a model in which net edge extension occurs by nascent focal adhesions advancing the site at which new actin arcs slow down and thus where the next protrusion phase begins (see model in Fig. 8). This would advance the base of the protrusion/retraction cycle in crawling cells. In non-crawling cells, the protrusion/retraction cycle of edge motion and its associated changes in actin dynamics are similar to crawling cells but the actin arcs cause nascent adhesions to slip further rearward during edge retraction. This would result in little or no advance of the base of the protrusion/retraction cycle. The system thus behaves like a mechanical ratchet with the actin arc acting as the lever and focal adhesions acting as the teeth. The extent of slippage of focal adhesions backward, dictated by the strength of an adhesion with the substrate, determines how quickly the cell advances forward. Within this system, the lamella would stay roughly the same width but, on the whole, advance due to new actin arc addition to the front of the lamella being balanced by removal of older actin arcs at the back of the lamella, as in a treadmill.
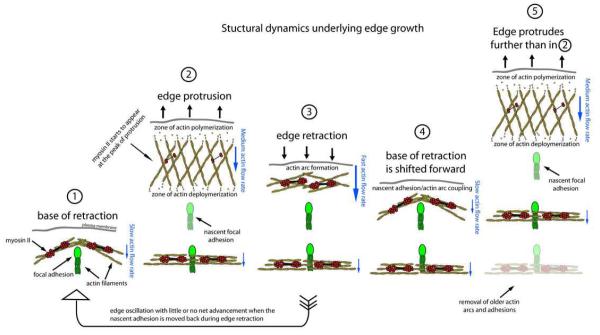
Figure 8.
Model of the structural dynamics of the actin cytoskeleton underlying edge motion. Leading edge advance is broken down into discrete steps. Step (1) shows the base of a previous retraction where a newly created actin arc is coupled to a focal adhesion. Hypothetically, the new lamellipodial protrusion could push off the arc to drive the membrane forward. During protrusion (2), actin filament polymerization occurs behind the plasma membrane and depolymerization occurs a few microns away from the edge. Actin filaments treadmill through the lamellipodium during protrusion, and nascent adhesions form. At the peak of protrusion (2) myosin II filaments form in the lamellipodium and a local network contraction (similar to that proposed for keratocyte cell body translocation17) occurs which drives actin arc formation and edge retraction (3). In cells that show net advance, the new actin arc slows at the nascent adhesion (4) due to most likely to strong coupling between the arc, adhesion, and growth substrate. The base of the retraction in (4) is shifted forward compared to (1). As a consequence, the start of the new protrusion in (5) is also shifted forward and the edge protrudes farther than in (2). In cells that do not show net advance, the actin arc and adhesion slip rearward during edge retraction (arrow from (3) to (1)). This indicates that there is still strong coupling between the actin arc and the adhesion, and also indicates a weak coupling between the adhesion and the growth substrate. Actin arc addition to the front of the lamella is balanced by actin arc removal at the back of the lamella (5). Lamellipodial and arc actin filaments are yellow. Focal adhesions and associated actin filaments are green. Myosin II filaments are red. Relative actin rearward flow rates are represented by blue arrows.
A cyclic myosin-II based actin arc formation and dissolution mechanism may also underlie the motile behavior in a variety of cells since most cells exhibit oscillatory edge protrusion and retraction. Indeed other motile cells have both a protrusion/retraction cycle and actin arcs. The list includes but is not limited to fish keratocytes, neuronal growth cones, and mouse melanoma cells (see references17, 27, 33 , Fig. S4 and Movie S4). Our findings of leading edge motion involving myosin II-based actin arc formation and dissolution thus provides a predictive model for cell crawling testable in a range of cell types.
Materials and Methods
Cell Culture and Chemicals
PtK1 cells were purchased from ATCC (Manassas, VA) and cultured in DMEM F12 (Invitrogen, Carlsbad, CA) as previously described5. Cells were plated on coverslips coated with 10μg/mL fibronectin for 2 hours at 37°C and cultured overnight. Cells were transfected with DNA plasmids with fugene (Roche, Indianapolis, IN) following manufacturer recommendations, and allowed to express for 12 hours. Cells were then imaged in CO2 independent media (Invitrogen) at 37°C. Blebbistatin, taxol, phalloidin, and fibronectin were from Sigma (Saint Louis, MO).
Actin Speckle Tracking
Time-lapse images of actin speckles were acquired with both widefield epifluorescence and spinning disk microscopy at intervals of 5 seconds with an integration time of 300-600ms with an Ultraview spinning disk confocal (Perkin Elmer, Waltham, MA) attached to an Olympus IX71 microscope (Olympus, Center Valley, PA) or a Marianas spinning disk (Intelligent Imaging Innovations, Denver, CO) attached to a Zeiss Observer.Z1 microcope (Carl Zeiss, Thornwood, NY). Actin speckles were generated by selective conversion of a sub-population of actin-tdEOS molecules by a 100-400ms exposure with 405nm laser light, through the spinning disk or by use of a Mosaic Digital illumination System (Photonic Instruments, Saint Charles, IL). An adaptive multi-frame correlation approach, as previously described23, was used to track actin-tdEOS speckles.
Photoconversion actin-tdEOS in specific cellular regions
Actin-tdEOS photoconversion experiments presented in figure 2 were preformed on a Marianas spinning disk equipped with a Mosaic digital illumination system. The laser power entering the Mosaic was 9 mW. Image acquisition and photoconversion of actin-tdEOS molecules (including region selection and 405 laser exposure control) was performed using Slidebook 5.0 software. All other experiments were performed with actin fused to monomeric fluorescent proteins. The incorporation of actin constructs into filaments in every observable actin-based structure was confirmed with live-cell extraction followed by a comparison with fluorescent phalloidin labeling.
Electron Microscopy
Electron microscopy of rotary shadowed cells was performed as previously described34. Briefly, cells were extracted with 1% Triton X-100 in cytoskeleton stabilization buffer (100mM PIPES, pH 6.9, 4% PEG, 10μM phalloidin, 10μM Taxol, 5mM EGTA, and 5mM MgCl2) for 5 minutes at room temperature. Cells were then washed with wash buffer 100mM PIPES, pH 6.9, 10μM phalloidin, 10μM Taxol, 5mM EGTA, and 5mM (MgCl2) for 2 minutes and fixed with 2% glutaraldehyde for 20 minutes. Fixation with 0.1% tannic acid (20 min) was followed by 0.2% uranyl acid (20 min). Samples were dehydrated with increasing concentrations of ethanol (10%, 30%, 50%, 70%, 90%, 100%, 100%, 100%, and 100% dried with molecular sieves purchased from Sigma) for 5 minutes each. Cells were critical point dried and rotary shadowed with platinum/carbon. All steps were done at room temperature. Images were acquired at 80Kv.
Analysis
Time-lapse images of actin-RFP were acquired at intervals of 10-20 seconds with an Ultraview spinning disk or a Marinas spinning disk microscope. Leading edge protrusion and retraction velocities were measured as previously described8, 16. This software package defines a spline representation of the leading edge and calculates edge displacements between frames. The spacing between displacement measurements along the edge was set at 2 pixels. Custom software was developed using Matlab (The Mathworks Inc, Natick, MA) to convert displacement measurements to velocities in nm/sec. The edge was divided into consecutive 172nm wide positions and velocities for each position over time were binned. For each cell, two matrices were constructed which contained the edge displacements and velocities, respectively, along different spatial positions (each matrix had N columns corresponding to N spatial positions interrogated). In the displacement matrix, each column represented a separate spatial position and contained the edge displacement values of that position at different time points. Similarly, each column in the velocity matrix contained the velocity values corresponding to the evolution of velocity over time at that particular spatial position.
Measurement of Protrusion/Retraction Period
The primary frequency of velocity fluctuation at each spatial position (bin) was identified by Fourier transforming the velocity values drawn from the corresponding column of the velocity matrix, and then the time-period was subsequently calculated using this primary frequency. Finally, the characteristic frequency and time period of protrusion/retraction of the cell were calculated by averaging the primary frequencies and time-periods, respectively, of all the spatial positions.
Protrusion Amplitude Measurements
For each cell, the time points for the start and end of each protrusion was initially calculated using the velocity matrix (constructed as described in previous section). The change in sign of velocity values were used to identify the start and end of protrusions, with the velocity values changing from negative to positive at the start of a protrusion (and end of preceding retraction), and, changing from positive to negative at the end of the protrusion (and start of the next retraction). In order to discard the extraneous cycles arising out of local fluctuations of velocity values during the transition from protrusion to retraction (and vice-versa), the cycles of protrusion/retraction with time-period less than half of the time-period (for that particular cell) identified by Fourier transformation (described in the previous section) were excluded.
Once the time points of start and end of protrusions were identified, the amplitude of each individual protrusion was calculated from the differences in displacements of each spatial position between the end and start of the protrusion. The retraction amplitudes were similarly calculated by finding out the difference between the end and start of the retractions (represented by the start of protrusion and the end of the previous protrusion). The average protrusion and retraction amplitudes of edge motion for each cell were obtained by averaging the corresponding values for all the spatial positions.
Measurement of Edge Growth
Finally the net edge growth for each cell was calculated from the differences in displacements of each spatial position between the position of the base of the first protrusion and the position of the base of the last protrusion. The edge growth rate in microns/minute could subsequently be calculated by dividing the net edge growth by the time interval between the two spatial positions.
Focal Adhesion Movement
Centroids of focal adhesions either labeled with zyxin-mCherry or vincullin-mCherry were tracked using the manual tracking protocol included in Slidebook. Next, a custom-written matlab code was used to measure the displacement of the newly formed focal adhesions over two protrusion/retraction cycles from their first point of appearance. Briefly, the protrusion or retraction cycle during which the focal adhesions first appeared was identified using the start/end points of the protrusion/retraction cycles obtained as described previously, and position of the closest preexisting focal adhesion for every new focal adhesion was determined. Finally, the displacement and the net displacement (displacement from the closest pre-existing focal adhesion) of each newly formed focal adhesion was calculated by tracking their spatial evolution during two consecutive protrusion/retraction cycles following their appearance.
Supplementary Material
Figure S1: Time montage showing a secondary actin bundle left behind by the protruding lamellipodium (yellow arrowhead). This bundle merges with the newly formed actin arc (red arrowhead) during edge retraction.
Figure S2: Focal adhesion slippage occurs during edge retraction. a) Schematic showing what time segmentation of focal adhesion movement quantified in b) and c). b) Total focal adhesion translocation during the protrusion phase of edge motion. c) Total focal adhesion translocation during the retraction phase of edge motion.
Figure S3: Actin arc coupling to nascent adhesions advances the edge’s protrusion retraction cycle. a) Time montage of paxllin-mCherry and actin-mGFP over three protrusion retraction cycles. Yellow arrowheads denote nascent adhesions and white arrowheads denote actin arc retraction between nascent adhesions. b-c) Paxillin (green) and actin (red) kymographs from line 1 (b) and line 2 (c) in (A). Line 1 is adjacent to the nascent adhesion formation and line 2 is a region directly on the adhesion. The number of protrusion/retraction cycles are labeled in (b). Arrowheads denote nascent adhesion formation. (c) Yellow arrowhead denotes rearward movement of the nascent adhesion and blue arrowhead denotes maturation. Red arrowheads denote the advance of the location of the base of the protrusion/retraction cycle after adhesion maturation. d) Time-montage of the box in (a). White arrowheads denote the appearance, yellow arrowheads denote the rearward movement, and blue arrows denote the maturation of adhesion.
Figure S4: Oscillatory edge motion at the leading edge of migrating fish keratocyes. Fish keratocytes have long been a favored model for cell migration because of their extremely fast migration rates. Although, electron microscopy studies have shown keratocytes have prominent myosin II-laden actin arcs, it has been assumed that their edge only protrudes, and, thus, does not display oscillatory motion 1, 2. We found that the published data was not sufficient to distinguish whether or not fish keratocytes had oscillatory edge motion. Therefore, we cultured keratocytes as previously described3 from the scales of Poecilia latipinna. We then acquired differential interference contrast time-lapse recordings of single crawling cells with images acquired every second. a) shows one frame of a time-lapse DIC recording of one of these crawling cells (dotted line shows orientation of kymographs). b-c) Kymographs are presented for three cells that are moving quickly (b) and slowly (c) relative to each other. All cells analyzed show edge oscillations (arrowheads). These oscillations do not always, but indeed can, result from an edge retraction, but, instead, a slowing of the edge protrusion. Cell body and cell velocity are indicated in the kymograph for each cell. Actin arc formation has been shown to be cyclical and myosin II-dependent in the neuronal growth cone with little, if any edge oscillations. It is important to notes that these studies were done using growth cones on poly-D-lysine coated coverslips on which growth cones do not extend. In contrast, neuronal growth cones plated on permissive growth substrates do show a robust edge oscillatory cycle identical to the motion presented here. The similarity in actin filament organization between the keratocyte1, neuronal growth cone4,5, and Ptk1 cells (Fig. 1) gives further support that these cells could use the same mechanism for motility.
Movie S1: Raw time-lapse recording of actin-tdEOS molecules at the leading edge. Epi-fluorescence. Images acquired every 5 seconds. Movie height: 33 mm. Movie length: 15.3 min.
Movie S2: Time-lapse recording of actin-RFP expressed in a PtK1 cell. Spinning disk confocal. Images acquired every 20 seconds. Movie height: 39 μm. Movie length: 60 min.
Movie S3: Time-lapse recording of actin-RFP (red) and myosin IIA-GFP (green) at the leading edge. Myosin IIA-GFP channel intensity was stretched to reveal the small amount of myosin in the lamellipodium. Spinning disk confocal. Images acquired every 10 seconds. Movie height: 31 μm. Movie length: 25.8 min.
Movie S4: Time-lapse recording of a mouse melanoma cell (B16-F1) expressing actin-mRFP. Note the cycle of actin arc assembly and disassembly underlying edge motion. It has been shown that actin filaments from the filopodial actin bundles (see prominent actin bundles a the leading edge) can be added to the actin arc bundles6. How this modification changes edge growth will be an interesting direction of future investigation. We focused our current study on actin arcs in cells that lack filopodial actin bundles for several reasons. First, actin arc assembly occurs in cells lacking filopodia1. Secondly, it has been shown that actin arc formation is unimpeded by filopodial actin bundle removal4. Taken together, this would indicate that the basal mechanism of actin arc format ion is independent of filopodia. B16-F1 cells were cultured as previously describe6. Images were acquired every 5 seconds. Movie height: 61 μm. Movie length: 14 min.
References
- 1.Rafelski SM, Theriot JA. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu Rev Biochem. 2004;73:209–39. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- 2.Mogilner A, Keren K. The shape of motile cells. Curr Biol. 2009;19:R762–71. doi: 10.1016/j.cub.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heath JP. Arcs: curved microfilament bundles beneath the dorsal surface of the leading lamellae of moving chick embryo fibroblasts. Cell Biol Int Rep. 1981;5:975–80. doi: 10.1016/0309-1651(81)90214-9. [DOI] [PubMed] [Google Scholar]
- 5.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–74. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Koestler SA, Auinger S, Vinzenz M, Rottner K, Small JV. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat Cell Biol. 2008;10:306–13. doi: 10.1038/ncb1692. [DOI] [PubMed] [Google Scholar]
- 7.Giannone G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 8.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–6. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 9.Giannone G, et al. Lamellipodial Actin Mechanically Links Myosin Activity with Adhesion-Site Formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small JV, et al. Unravelling the structure of the lamellipodium. J Microsc. 2008;231:479–85. doi: 10.1111/j.1365-2818.2008.02060.x. [DOI] [PubMed] [Google Scholar]
- 11.Vallotton P, Small JV. Shifting views on the leading role of the lamellipodium in cell migration: speckle tracking revisited. J Cell Sci. 2009;122:1955–8. doi: 10.1242/jcs.042036. [DOI] [PubMed] [Google Scholar]
- 12.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 13.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–94. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danuser G. Coupling the dynamics of two actin networks--new views on the mechanics of cell protrusion. Biochem Soc Trans. 2005;33:1250–3. doi: 10.1042/BST0331250. [DOI] [PubMed] [Google Scholar]
- 15.Danuser G. Testing the lamella hypothesis: the next steps on the agenda. J Cell Sci. 2009;122:1959–62. doi: 10.1242/jcs.054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–52. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–30. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XF, Schaefer AW, Burnette DT, Schoonderwoert VT, Forscher P. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 2003;40:931–44. doi: 10.1016/s0896-6273(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 20.Jurado C, Haserick JR, Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol Biol Cell. 2005;16:507–18. doi: 10.1091/mbc.E04-10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–51. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–35. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 23.Ji L, Danuser G. Tracking quasi-stationary flow of weak fluorescent signals by adaptive multi-frame correlation. J Microsc. 2005;220:150–67. doi: 10.1111/j.1365-2818.2005.01522.x. [DOI] [PubMed] [Google Scholar]
- 24.Welch MD, Mallavarapu A, Rosenblatt J, Mitchison TJ. Actin dynamics in vivo. Curr Opin Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- 25.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–5. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 26.Gardel ML, et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–26. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 28.Choi CK, et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–50. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrova AY, et al. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS One. 2008;3:e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aratyn-Schaus Y, Gardel ML. Transient frictional slip between integrin and the ECM in focal adhesions under myosin II tension. Curr Biol. 2010;20:1145–53. doi: 10.1016/j.cub.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shemesh T, Verkhovsky AB, Svitkina TM, Bershadsky AD, Kozlov MM. Role of focal adhesions and mechanical stresses in the formation and progression of the lamellipodium-lamellum interface [corrected] Biophys J. 2009;97:1254–64. doi: 10.1016/j.bpj.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossier OM, et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. Embo J. 2010;29:1055–68. doi: 10.1038/emboj.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mongiu AK, Weitzke EL, Chaga OY, Borisy GG. Kinetic-structural analysis of neuronal growth cone veil motility. J Cell Sci. 2007;120:1113–25. doi: 10.1242/jcs.03384. [DOI] [PubMed] [Google Scholar]
- 34.Burnette DT, Schaefer AW, Ji L, Danuser G, Forscher P. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat Cell Biol. 2007;9:1360–9. doi: 10.1038/ncb1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Time montage showing a secondary actin bundle left behind by the protruding lamellipodium (yellow arrowhead). This bundle merges with the newly formed actin arc (red arrowhead) during edge retraction.
Figure S2: Focal adhesion slippage occurs during edge retraction. a) Schematic showing what time segmentation of focal adhesion movement quantified in b) and c). b) Total focal adhesion translocation during the protrusion phase of edge motion. c) Total focal adhesion translocation during the retraction phase of edge motion.
Figure S3: Actin arc coupling to nascent adhesions advances the edge’s protrusion retraction cycle. a) Time montage of paxllin-mCherry and actin-mGFP over three protrusion retraction cycles. Yellow arrowheads denote nascent adhesions and white arrowheads denote actin arc retraction between nascent adhesions. b-c) Paxillin (green) and actin (red) kymographs from line 1 (b) and line 2 (c) in (A). Line 1 is adjacent to the nascent adhesion formation and line 2 is a region directly on the adhesion. The number of protrusion/retraction cycles are labeled in (b). Arrowheads denote nascent adhesion formation. (c) Yellow arrowhead denotes rearward movement of the nascent adhesion and blue arrowhead denotes maturation. Red arrowheads denote the advance of the location of the base of the protrusion/retraction cycle after adhesion maturation. d) Time-montage of the box in (a). White arrowheads denote the appearance, yellow arrowheads denote the rearward movement, and blue arrows denote the maturation of adhesion.
Figure S4: Oscillatory edge motion at the leading edge of migrating fish keratocyes. Fish keratocytes have long been a favored model for cell migration because of their extremely fast migration rates. Although, electron microscopy studies have shown keratocytes have prominent myosin II-laden actin arcs, it has been assumed that their edge only protrudes, and, thus, does not display oscillatory motion 1, 2. We found that the published data was not sufficient to distinguish whether or not fish keratocytes had oscillatory edge motion. Therefore, we cultured keratocytes as previously described3 from the scales of Poecilia latipinna. We then acquired differential interference contrast time-lapse recordings of single crawling cells with images acquired every second. a) shows one frame of a time-lapse DIC recording of one of these crawling cells (dotted line shows orientation of kymographs). b-c) Kymographs are presented for three cells that are moving quickly (b) and slowly (c) relative to each other. All cells analyzed show edge oscillations (arrowheads). These oscillations do not always, but indeed can, result from an edge retraction, but, instead, a slowing of the edge protrusion. Cell body and cell velocity are indicated in the kymograph for each cell. Actin arc formation has been shown to be cyclical and myosin II-dependent in the neuronal growth cone with little, if any edge oscillations. It is important to notes that these studies were done using growth cones on poly-D-lysine coated coverslips on which growth cones do not extend. In contrast, neuronal growth cones plated on permissive growth substrates do show a robust edge oscillatory cycle identical to the motion presented here. The similarity in actin filament organization between the keratocyte1, neuronal growth cone4,5, and Ptk1 cells (Fig. 1) gives further support that these cells could use the same mechanism for motility.
Movie S1: Raw time-lapse recording of actin-tdEOS molecules at the leading edge. Epi-fluorescence. Images acquired every 5 seconds. Movie height: 33 mm. Movie length: 15.3 min.
Movie S2: Time-lapse recording of actin-RFP expressed in a PtK1 cell. Spinning disk confocal. Images acquired every 20 seconds. Movie height: 39 μm. Movie length: 60 min.
Movie S3: Time-lapse recording of actin-RFP (red) and myosin IIA-GFP (green) at the leading edge. Myosin IIA-GFP channel intensity was stretched to reveal the small amount of myosin in the lamellipodium. Spinning disk confocal. Images acquired every 10 seconds. Movie height: 31 μm. Movie length: 25.8 min.
Movie S4: Time-lapse recording of a mouse melanoma cell (B16-F1) expressing actin-mRFP. Note the cycle of actin arc assembly and disassembly underlying edge motion. It has been shown that actin filaments from the filopodial actin bundles (see prominent actin bundles a the leading edge) can be added to the actin arc bundles6. How this modification changes edge growth will be an interesting direction of future investigation. We focused our current study on actin arcs in cells that lack filopodial actin bundles for several reasons. First, actin arc assembly occurs in cells lacking filopodia1. Secondly, it has been shown that actin arc formation is unimpeded by filopodial actin bundle removal4. Taken together, this would indicate that the basal mechanism of actin arc format ion is independent of filopodia. B16-F1 cells were cultured as previously describe6. Images were acquired every 5 seconds. Movie height: 61 μm. Movie length: 14 min.