Figure 5.
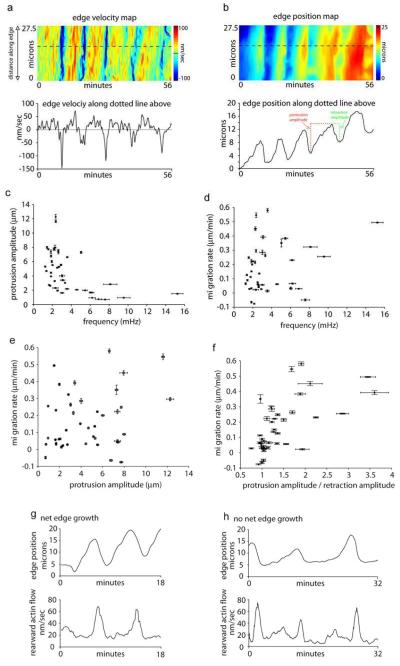
Oscillatory edge motion and net edge extension. a) Edge velocity map along the edge and a single region (bottom graph) along the edge from a crawling cell. b) Edge position map of the same cell as in (a). Edge position map was created by color-coded the lowest edge position as blue and the highest as red as in the color bar. This allows for relative edge position along the same regions as in the velocity map in (a) to be graphically displayed over time. Bottom graph in (b) shows the relative edge position at one point along the edge. Red dotted line and green dotted line graphically show the protrusion amplitude and retraction amplitude of one protrusion respectively. c) Protrusion amplitudes plotted against edge oscillation frequencies for individual cells. Correlation coefficient: −0.5129 (confidence interval: −0.7087, −0.2437). d) Migration rate plotted edge oscillation frequencies for individual cells. Correlation coefficient: 0.1966 (confidence interval: −0.1182, 0.4755). e) Migration rate plotted against protrusion amplitudes for individual cells. Correlation coefficient: 0.2650. (confidence interval: −0.0464, 0.5295). f) Migration rate plotted against the ratio of protrusion and retraction amplitudes in individual cells. Correlation coefficient: 0.4254 (confidence interval: 0.1355, 0.6482). g-h) Edge position and rearward flow velocity plotted as in Fig. 1(f) for a cell that demonstrates net edge growth (g) and a cell that does not (h). Note the pattern of rearward actin flow is similar in both cells. Pearson’s correlation coefficient was used to quantify the correlation and the 95% confidence interval for each pair was computed by using the Fisher transformation.