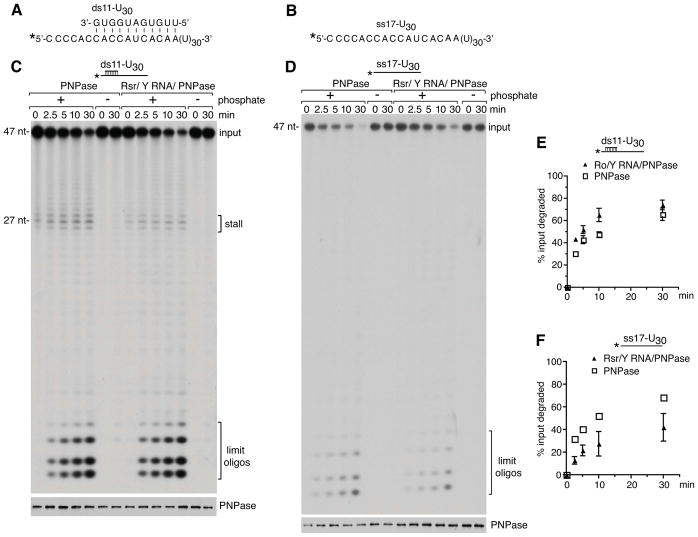
Figure 5. The RNP degrades a duplex more effectively than PNPase but is less active on single-stranded RNA.
(A and B) Sequences of ds11-U30 and ss17-U30 substrates. The ss17-U30 RNA corresponds to the overhang strand of the ds11-U30 duplex. In both substrates the 5′ end of the ss17-U30 RNA was labeled with [γ-32P]ATP.
(C and D) The activity of PNPase and the RNP was compared on the ds11-U30 (C) and ss17-U30 (D) substrates. Aliquots were removed at intervals and fractionated in denaturing gels to visualize the labeled RNA (top panels). Aliquots were also assayed by immunoblotting to confirm that equal amounts of PNPase were present (bottom panels). Notably, although more of the RNP may have been used in the ss17-U30 assay (D, bottom panel), the RNP is less active than PNPase on this substrate (top panel).
(E and F) Degradation reactions using the ds11-U30 and ss17-U30 substrates were performed in triplicate. Data are represented as mean values from three replicates ± SEM. In (F), error bars for PNPase are too small to be visible.
See also Figure S4.