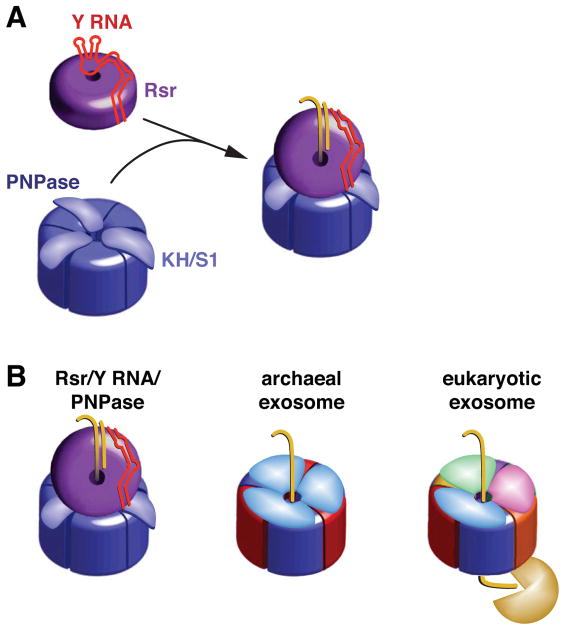
Figure 7. Model for Rsr/Y RNA/PNPase RNP function.
(A) Formation of the Rsr/Y RNA/PNPase RNP. Residues on the Rsr outer surface contact conserved bases in the Y RNA stem, while the rest of the RNA interacts with basic residues on the Rsr surface, preventing binding of other RNAs (Stein et al., 2005). When PNPase is present, Y RNA loops bind the PNPase KH and S1 domains, repositioning the RNA so that substrates can access the Rsr cavity.
(B) The Rsr/Y RNA/PNPase RNP resembles archaeal and eukaryotic exosomes. All three complexes contain rings with six RNase PH domains. PNPase is a trimer in which each monomer contains two RNase PH domains. The archaeal ring contains three copies of each of two proteins containing single RNase PH domains, while in eukaryotes six distinct proteins form the ring. Both exosomes contain RNA-binding caps formed by KH and S1 domain proteins. We propose that, similar to the archaeal exosome, RNA passes through an RNA-binding ring (Rsr) to undergo phosphorolytic degradation in the catalytic ring (PNPase). As yeast and human exosomes have a catalytically inactive RNase PH domain ring, RNA passes through the nine subunit Exo1–9 ring to reach a hydrolytic exonuclease.