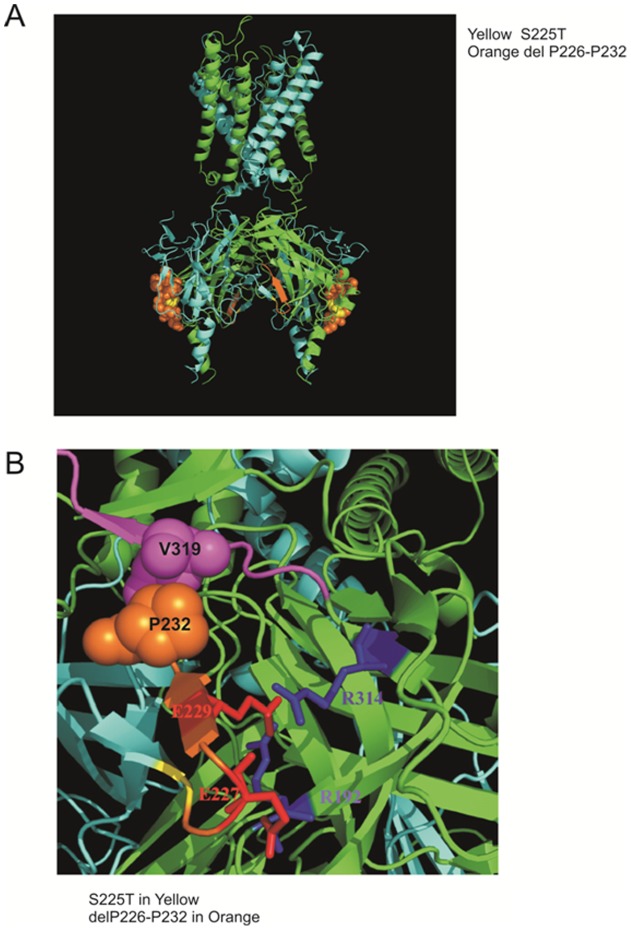
Figure 5. Homology modeling of Kir6.2 from Kir2.2 structure with PyMOL software.
(A) S225T is colored in yellow and deleted amino acids 226–232 (-PEGEVVP-) are colored in orange. (B) E227 and E229 are colored and labeled in red while R192 and R314 are colored and labeled in blue. The structure model reveals the possible interaction between the deleted amino acid P232 (orange spheres) and V319 in the proposed Kir6.2 Ankyrin-B binding site (a.a. 316 to a.a. 323 –VPIVAEED- colored in magenta).