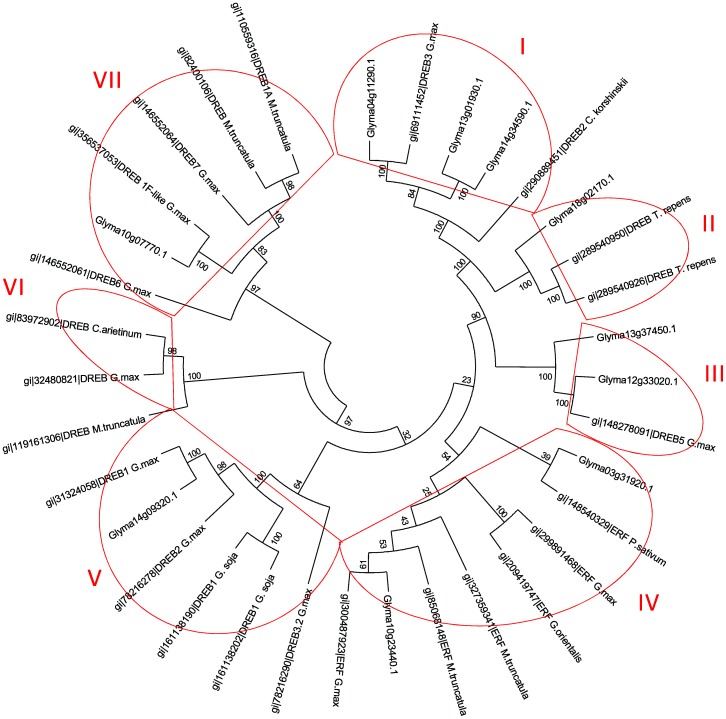
Figure 2. Phylogenetic tree. Proteins encoded by the candidate genes and the DREB/ERF protein that was described in the NCBI database were used to construct the tree using the ClustalW algorithm with the MEGA 5 program.
The Neighbor-Joining (NJ) method was used with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed). Candidate genes are represented by the GeneModels, and the homologous DREB/ERF sequences from Fabaceae (Glycine max, Medicago truncatula, Cypripedium arietinum, Trifolium repens, Glycine soja, Caragana korshinskii, Pisum sativum, and Galega orientalis) are represented by GI.