Table 2. Optimal segmentation for data from yeast.
| Network property | Type | k | Segments |
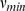
|
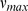
|
| relative density | G | 6 | [1]–[4],[5]–[9],[10]–[13],[14]–[20],[21]–[31],[32]–[36] | 0.05 | 5.60 |
| degree | L | 8 | [1]–[4],[5]–[8],[9]–[12],[13]–[16],[17]–[20],[21]–[24],[25]–[32],[33]–[36] | 4.35 | 13.50 |
| closeness | LG | 6 | [1]–[4],[5]–[9],[10]–[17],[18]–[21],[22]–[31],[32]–[36] | 0.05 | 4.24 |
| betweenness | LG | 6 | [1]–[4],[5]–[8],[9]–[12],[13]–[20],[21]–[24],[25]–[36] | 3.80 | 14.52 |
| Existing method | k | Segments |
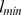
|
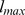
|
|
| Ramakrishnan et al. [15] | 8 | [1]–[6],[7]–[10],[11]–[14],[15]–[18],[19]–[22],[23]–[26],[27]–[31],[32]–[36] | 4 | 7 |
The upper part of the table shows the result of the optimal segmentation for synthetic data based on dynamic programming, while the lower part contains the result based on the method of Ramakrishnan et al.
[15]. In the upper table, the first and second columns show the name and the type of network properties used to determine the distances: G stands for global, L for local, and LG for local-global. The third column includes the number 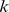 of segments that maximize the objective
of segments that maximize the objective 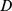 with the dynamic programming approach. The resulting segments are given in the forth column, while the fifth and sixth columns contain the corresponding values of lower (
with the dynamic programming approach. The resulting segments are given in the forth column, while the fifth and sixth columns contain the corresponding values of lower (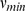 ) and upper (
) and upper (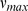 ) bound of the tuning parameter
) bound of the tuning parameter  . The lower part also includes minimum and maximum length of the segments, i.e.,
. The lower part also includes minimum and maximum length of the segments, i.e., 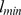 and
and 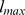 , as parameters of the contending method.
, as parameters of the contending method.
