Figure 4. IL-22 deficient and co-housed wild-type mice have altered microbiota compared to wild-type mice by 16S rRNA analysis of fecal-derived bacterial microbiota.
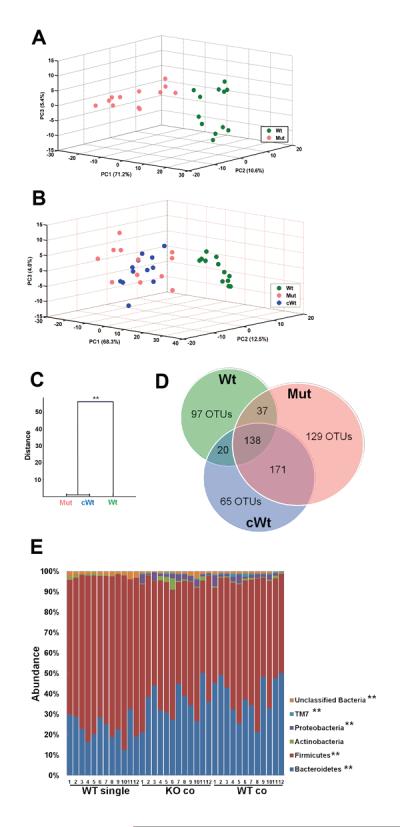
Prior to DSS-treatment, fecal samples were obtained from the mice described in Figure 2. DNA was prepared and then subjected to pyrosequencing. 68,822 total sequences were obtained from the 36 samples (n=12/group). (A) Score plot of PCA of the microbiomes of wild-type (Wt) and IL-22 deficient mice (Mut) based on the first three PCs. Each dot represents one mouse. (B) Score plot of PCA based on the first three PCs of wild-type mice (Wt), IL-22 deficient mice (Mut) and wild-type mice housed with IL-22 deficient mice (cWt). Each dot indicates one mouse. (C) Clustering of bacteria based on distances between different groups calculated with multivariate analysis of variance test of the first seven PCs of UniFrac PCoA analysis data. The Mahalanobis distances between group means are shown. ** p<0.01 (D) Venn diagram showing distribution of the shared OTUs. (E) Taxon-based analysis at phylum level among the groups. Phyla above 1% abundance are shown (Kruskal-Wallis test, **p<0.01, *p<0.05).
