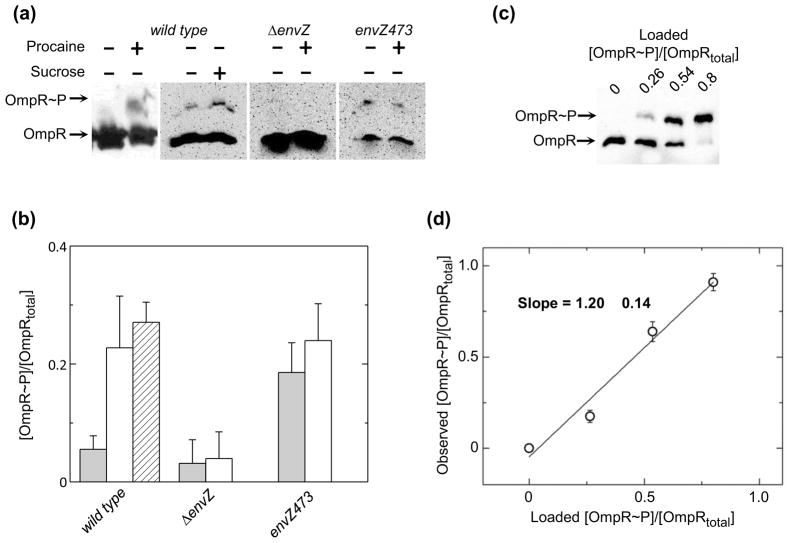
Fig. 2.
Characterization of OmpR phosphorylation in E. coli cells. (a) Representative western blots of phosphoprotein affinity gel electrophoretic separation of wild type, ΔenvZ and envZ473 mutant cell lysates in the presence and absence of procaine or sucrose, as indicated. Representative blots are taken from separate phosphoaffinity SDS-PAGE gels. Therefore, differences in signal intensity do not reflect differences in cellular OmpR concentration. (b) Quantitation of the extent of OmpR phosphorylation in wild type, ΔenvZ and envZ473 cell lysates, untreated (clear bars), following treatment with procaine (shaded bars), or following addition of sucrose to the media (hashed bar). Values reflect the mean values of 3 separate experiments with error bars reflecting the standard deviation from the mean. (c) Representative western blot of known fractions of [OmpR~P]/[OmpRtotal] mixed with ΔompR cells and subsequently lysed. (d) Quantitative efficiency of western blots of phosphoprotein affinity gel electrophoretic separations of [OmpR~P]/[OmpRtotal]. Points represent the average values from 2 separate experiments with error bars reflecting the standard deviation from the mean. Solid line indicates the best-fit linear regression analysis of the plotted data.