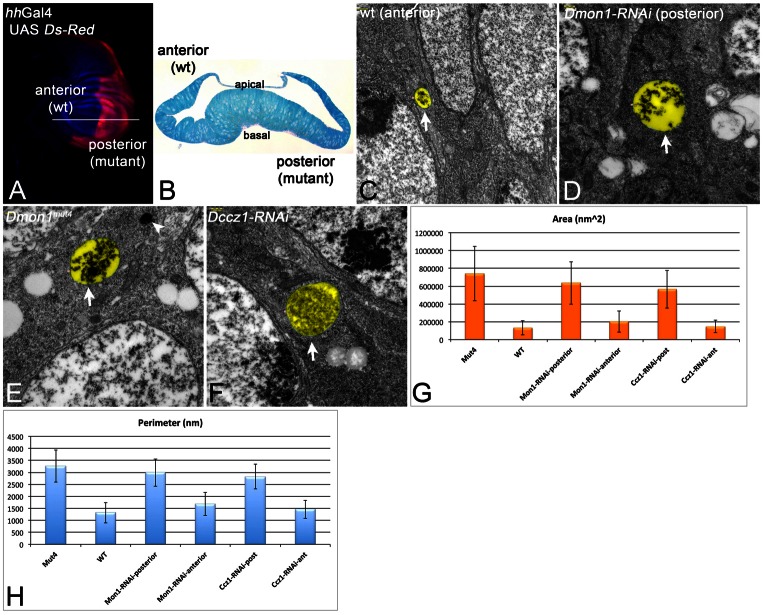
Fig. 6.
TEM analysis of Dmon1 and Dczz1 mutants. (A) Expression pattern of hhGal4 revealed by UAS dsRed. hhGal4 drives expression of UAS constructs in the posterior compartment. It was used to express the UAS RNAi constructs together with UAS Dcr2. The discs were sectioned at the level of the white line and the phenotypes of the posterior cells were compared to the anterior wild-type (wt) cells. (B) Semi cross-section at the level indicated by the white line in A. (C,D) Representative MVBs (arrows) of anterior wild-type (C) and posterior Dmon1-depleted (D) cells. (E) Representative MVB (arrow) of Dmon1mut4 cells. The MVBs of Dmon1 mutant cells are larger. The arrowhead in E points to a lysosome of normal appearance. (F) Representative MVB (arrow) of cells depleted of Dccz1 function. The magnification is the same in C–F. The MVBs are highlighted in pseudo-colour in C–F. (G,H) Statistical analysis of the area and perimeter of the MVBs underscores the enlargement of MVBs in mutant cells.