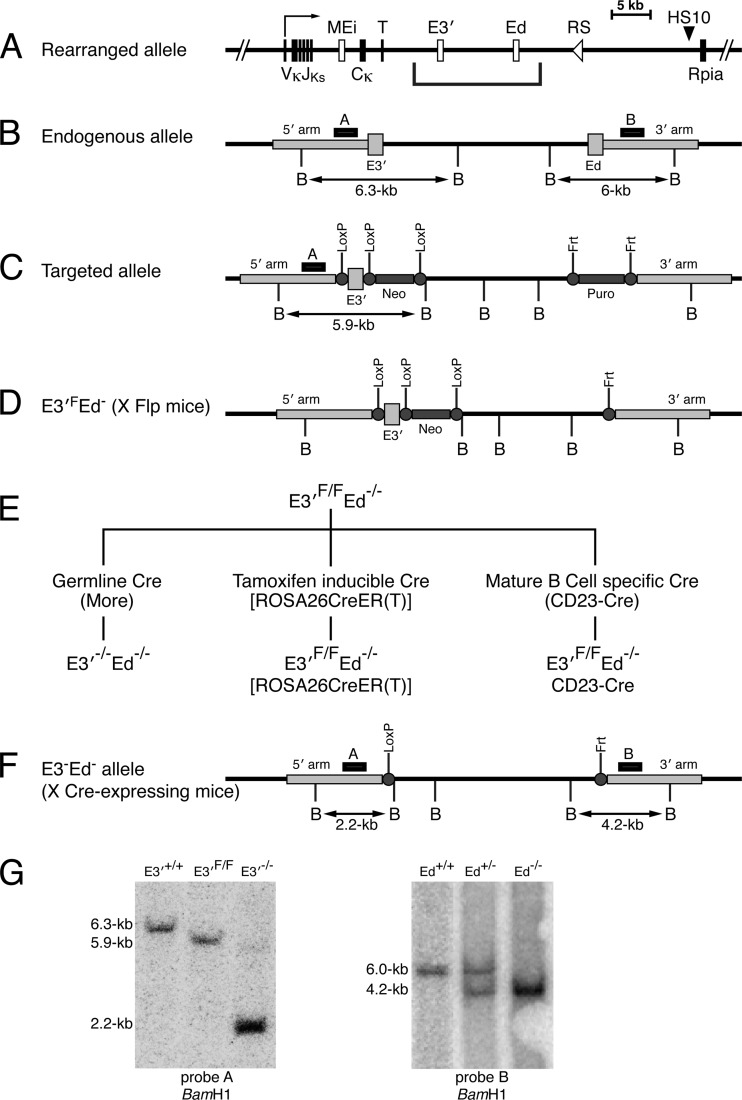
Fig 1.
Strategy for creating and testing conditional E3′−/− Ed−/− knockout mice. (A) Schematic diagram depicting a rearranged Igκ locus. The coordinates of Ei, E3′, Ed, and HS10 in the NCDI37/mm9 mouse chromosome 6 sequence are 70,675,570 to 70,676,084, 70,685,250 to 70,686,058, 70,693,704 to 70,694,944, and 70,713,758 to 70,715,084, respectively. MEi, T, and Rpia correspond to the matrix association region intronic enhancer element, the transcription termination region, and the ribose 5′-phosphate isomerase gene, respectively. The bracket below the schematic diagram demarcates the relevant region targeted in these studies. (B) Endogenous Igκ locus segment possessing E3′ and Ed. The positions of probes A and B are indicated by small black boxes above the map. BamHI restriction sites (B) are indicated below the map. (C) Targeted allele of the Igκ locus. The targeting construct was made using BAC recombineering as previously described (12). E3′ was replaced with a Neo+ E3′ cassette flanked by loxP sites, and Ed was replaced with a Puro gene flanked by Frt sites. (D) The Puro gene was deleted by breeding the mice with a Flp-expressing strain (28) to obtain E3′F Ed− mice. It should be emphasized that despite the fact that the Neo gene is still present adjacent to E3′ in this conditional allele, B cell development and transcription of rearranged Igκ genes are very similar to those in Ed−/− mice (see Fig. 2A, top panels). (E) Experimental strategy used to delete E3′ in the mouse germ line (left segment of the flow chart), in any cell type upon tamoxifen treatment (middle segment of the flow chart), or specifically in mature B cells (right segment of the flow chart). (F) Structure of the conditional allele after deletion of E3′ by expressing Cre recombinase. (G) Southern blotting of BamHI-digested mouse tail DNA. The mouse genotypes studied are indicated above the lanes. The locations of probes A and B are shown above the map in panel F. The left blot demonstrates as a control that the E3′F/F allele is successfully deleted upon breeding such mice with mice expressing Cre recombinase in the germ line (27). The right blot demonstrates that the Puro gene was successfully deleted upon breeding mice with mice expressing Frt recombinase in the germ line (28).